Comparison of responses by bacteriophage and bacteria to pressures on the base composition of open reading frames
J. R Mortimer and D. R. Forsdyke
Applied Bioinformatics (2003) 2, 47-62.
Copying of this article, including placing at another website, requires the written permission of Open Mind Journals, the copyright owner.
Keywords: Bacteriophage; base composition; Chargaff difference; genome phenotype; purine-loading
![]()
Purine-loading in phage lambda
Purine excess or pyrimidine deficiency?
Contributions of different codon positions to (G+C)%
Contributions of different codon positions to PLI
Inverse relationship between PLI and (G+C)%
W and S base contributions to PLIs
Quadrant plots
Contributions of codon positions to individual base pressures
Disparities in purine-loading between specific bacteriophages and their hostsPartial genome sequences
Translation-pressure
Purine-loading as a viral adaptation
Inverse relationship between purine-loading and (G+C)%
Relative strengths of A and G
Phage-host relationships
Conclusions
![]()
Abstract: Differences in genome base-composition can occur with respect to GC-pressure, purine-loading pressure (AG-pressure), and RNY-pressure, for which there are possible functional explanations, and with respect to the more abstract pressures exerted by individual bases. The graphical approach of Muto and Osawa was used to analyze how bacteriophages and bacteria balance potentially conflicting pressures on their genomes. Phages generally respond to AG-pressure by increasing A while keeping T constant, and by decreasing C while keeping G constant. In contrast, bacteria generally increase both A and T, the former more so, and decrease both G and C, the latter more so. These differences largely relate to third codon positions, which are more responsive than first and second codon positions to AG-pressure and GC-pressure. Phages respond to AG-pressure more in the third codon position than bacteria, whereas bacteria respond more in the first codon-position than phage. Conversely, bacteria respond to GC-pressure more in the third codon position than phages, whereas phages respond more in the first codon position than bacteria. As GC-pressure increases, A is traded for C and AG-pressure decreases; first and second codon positions, having more A than T, are most responsive to this negative effect of increased GC-pressure; third positions either do not respond (phage), or respond weakly (bacteria). In a set of 48 phage/host pairs, degrees of purine-loading were less correlated than GC percentages. These results suggest that pressures on conventional and genome phenotypes operate differentially in phage and bacteria generating both general differences in base composition, and specific differences characteristic of particular phage-host pairs. The reciprocal relationship between GC-pressure and AG-pressure implies that effects attributed to GC-pressure may actually be due to AG-pressure, and vice-versa. |
![]()
Genomes are channels carrying multiple forms of information through the generations from the distant past to the present. As with information channels in general, carrying capacity might be finite and messages subject to distortion and noise. If genomes were unable to satisfy all informational demands, a balance would have to have been established with trade-offs between competing demands. This contrasts with the view that there is an excess of carrying capacity in genome information space so that "neutral" mutations can exist, and sequences without obvious function can be dismissed as "junk" (Forsdyke 2002a; Forsdyke et al 2002). Thus, our tasks are to identify the forms of information that genomes convey, and show if and how the demands of different forms are balanced according to the needs of individuals, species and higher taxa (Brenner 1991). The most widely recognized form of genomic information is genic. Proteins and RNAs encoded by genes appear to have most influence on the conventional phenotype, - an individual's somatic form and function. However, other types of information exert pressures on genome space. These pressures sometimes influence the conventional phenotype, often influence base composition, and are either local or general. Local pressures include purine-loading pressure (AG-pressure), and RNY-pressure. General pressures affect entire genomes and include GC-pressure, fold pressure and pressures for genome compactness (i.e. there is a "genome phenotype"; Schaap 1971; Forsdyke and Mortimer 2000). In some circumstances pressures appear to conflict. Thus, seeming to accommodate to AG-pressure, the lengths of an intracellular pathogen's proteins increase by inclusion of low complexity, inter-domain, segments, that do not appear important for protein function, yet contain amino acids encoded by AG-rich codons (Pizzi and Frontali 2001; Cristillo et al. 2001; Forsdyke 2002a; Xue and Forsdyke 2003). Because they often share the same intracellular environment, the genomes of viruses and their hosts could have many features in common, and respond similarly to local and general pressures on genome space. Differential responses might reflect fundamental differences between the biology of virus and host, which include host defence strategies against viruses, or virus attack strategies against hosts (Forsdyke et al 2002). Knowledge of such differences should assist their exploitation for therapeutic purposes. Ultimately the individual base compositional characteristics of each virus-host pair must be described, and its adaptive value determined. However, first we should know whether there are general differences between the base compositions of viruses and their hosts. We begin here at the most elementary level, with the viruses that infect bacteria (Hedrix et al 1999), including some human pathogens (Summers 2001). We report base compositional differences between bacteriophages in general, and bacteria in general, with particular emphasis on those attributable to AG-pressure. |
|
Purine-loading in phage lambda The clustering of purines in mRNA synonymous strands, as noted for bacteriophage lambda (Szybalski et al 1966), is sufficient to affect base composition and is a general phenomenon found both in prokaryotes and eukaryotes (Smithies et al. 1981; Saul and Battistuta 1988; Forsdyke and Mortimer 2000). By counting bases in 1 kb moving windows one can determine for the "top" strand of uncharted DNA sequences, where ORFs (or regions transcribed but not translated; Schattner 2002) are likely to be located, their direction of transcription, and their relationship to the origin of replication. For ORFs transcribed to the right (where the "top" strand is the mRNA-synonymous strand), Chargaff differences (ΔW, ΔS; see Methods) tend to favour purines. For ORFs transcribed to the left (where the "bottom" strand is the mRNA-synonymous strand), Chargaff differences tend to favour pyrimidines. The origin of replication on a bidirectionally-replicating chromosome can be close to a region where transcription directions, and hence degrees of top-strand purine-loading, can switch (Szybalski et al 1969; Smithies et al 1981; Frank and Lobry 1999; Tillier and Collins 2000). |
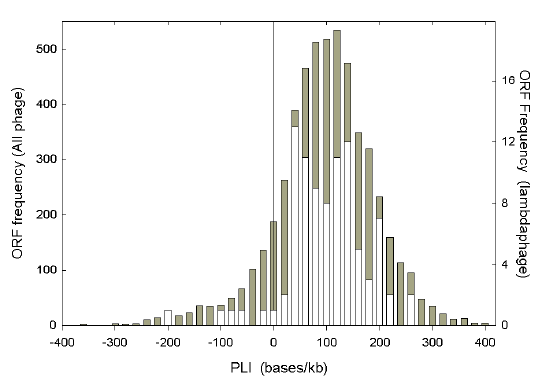 |
| Fig. 1. Distribution of Chargaff differences as assessed by the purine-loading index (PLI). Data are from 91 ORFs of lambdaphage (white, and right axis), and 5298 ORFs of all phage in the September 2000 release of GenBank (gray, and left axis). |
If transcription directions and limits of ORFs are known, Chargaff differences for the mRNA-synonymous strands of individual ORFs can be calculated directly from ORF base compositions. Values for ΔW and ΔS can be combined as the purine-loading index (PLI) for each ORF. Fig. 1 shows the frequency distribution of PLI values for 91 lambda phage ORFs (in white), against a background (in gray) of the 5298 ORFs from the 326 bacteriophage species in the September 2000 release of GenBank. The frequency distribution for lambda phage, while irregular, shows a spread similar to that of the general distribution, with most ORFs being purine-loaded. The average of the PLI values for the 5928 ORFs, some from species represented in GenBank by only one ORF, is 87.7 bases/kb. The average of the 91 lambda phage ORF PLI values is 88.8 bases/kb. When the bases of all lambda phage ORFs are pooled to calculate the average PLI for the genome (thus placing more emphasis on long ORFs) the value is 85.1 bases/kb. Some phage species have much higher, and some have much lower, values. These average genomic PLI values for each member of the subset of 161 phage species (from the above mentioned total of 326 species) with more than three available ORFs and 2500 bases were used in the studies that follow. Purine excess or pyrimidine deficiency? Purine-loading is evident when purines exceed pyrimidines, and a sequence where this holds can be considered to have responded to purine-loading-pressure (AG-pressure). Singly and collectively, bases contribute to (or detract from) purine-loading. The cause of the pressure to purine-load may be obvious (e.g. the need for amino acids with purine-rich codons in the highly conserved active site of an enzyme), or subtle (e.g. perhaps the need to avoid RNA-RNA interactions, which might generate double-stranded RNA; Forsdyke and Mortimer 2000; Forsdyke et al. 2002). |
 |
|
Fig. 2. Relative contributions of purine excess and pyrimidine deficiency to Chargaff differences. Data are for the coding regions of the 161 phage species (A, C), and the 1046 bacterial species (B, D), represented in GenBank by more than 3 ORFs and more than 2500 bases. Filled circles, A; open circles, T; filled squares, G; open squares, C. Parameters of the linear regression plots are: Y0, Y values when the PLI is zero; Sl, slopes; r2, adjusted coefficients of determination. Unless otherwise stated, the probabilities that slope values are not significantly different from zero are <0.0001. Note that when the PLI is zero, there is a slight A deficiency (259.0 - 272.5 = -13.5) and a slight G excess (+ 13.6) in phage, and a slight A excess (+3.2) and a slight G deficiency (-3.2) in bacteria. |
The relative contributions of the individual bases of each Chargaff pair to Chargaff differences are shown in Fig. 2, where each species is represented by a single point. As expected, linear regression plots cross when PLI values are near zero, and most species are to the right of zero (positive PLI values). As AG-pressure increases, phages increase ΔW values mainly by loading A, and, to a small extent, by unloading T (Fig. 2a). In contrast, bacteria load A, even more than phage, but this response to AG-pressure is offset by T-loading (Fig. 2b). Phages increase ΔS values mainly by unloading C, and, to a small extent, this is offset by some unloading of G (Fig. 2c). In contrast, bacteria unload G more than phage, and only achieve positive ΔS values by unloading more C than G (Fig. 2d).
Bases in third codon positions are usually least involved in determining an encoded amino acid (i.e. when mutated there is usually no change in the amino acid). As such, third bases can provide a guide to pressures on nucleic acids other than the pressures to encode specific proteins. For various bacterial species, Muto and Osawa (1987) plotted the average (G+C)% for each of the three codon positions against total genomic (G+C)%, and noted that the third position was the most responsive to changes in genome (G+C)% (i.e. the unit change in slope per unit change in GC% was greatest for this position). Least responsive was the second codon position, the position most involved in determining the encoded amino acid (i.e. when mutations occur here the encoded amino acid usually changes, and the organism may be negatively selected if the function of the corresponding protein is impaired). |
 |
| Fig. 3. Relative responses of different codon positions to GC-pressure (A, B) and AG-pressure (C, D), in bacteriophage (A, C) and bacteria (B, D). Open circles, first codon position; filled squares, second codon position; filled triangles, third codon position. Note that, for each set of three plots, the sum of Y0 values is zero. |
|
Figs. 3a,b show similar plots for the 161 bacteriophage species and the 1046 bacterial species represented in GenBank by more than three open reading frames (ORFs) and more than 2500 nucleotides. As the genomic (G+C)% of species increases, the (G+C)% at each codon position increases. Values for the first position range from 30-80%. Values for the second position range from 20-55%. Values for the third position range from 10% to nearly 100%. Thus, with respect to (G+C)%, the positions can be considered to follow an "SWN" rule, with first positions tending to be enriched for G and C (the S bases), and second positions tending to be enriched for A and T (the W bases). The (G+C)% of the third position increases from values initially below those of the first and second codon positions in species with low genomic (G+C)%, to become the predominant contributor to (G+C)% in species with high genomic (G+C)%. For all species, the first codon position always has a higher (G+C)% than the second codon position, and responds more to GC-pressure than the second codon position (greater slope P<0.0001). Although, as in other taxa, the second position responds the least, it does respond significantly, and this, together with the response of the first position can influence the average amino acid content of the encoded proteins (i.e. species with a high (G+C)% have more amino acids corresponding to GC-rich codons; Sueoka 1961; Knight et al 2001). In general, bacteriophage species are slightly more responsive to GC-pressure in the first and second codon positions than bacteria [slopes of 0.73 versus 0.69 (P=0.06), and 0.52 versus 0.43 (P=0.0002), respectively]. However, bacteria are more responsive to GC-pressure in the third codon position (slope of 1.88 versus 1.75; P=0.0001; Figs. 3a,b).
Contributions of different codon positions to PLI The responsiveness of different codon positions to AG-pressure is shown in Figs. 3c,d. In keeping with the "RNY rule" (Eigen and Schuster 1978; Shepherd 1981; Kypr and Mrazek 1987), for all species the first codon position makes the greatest contribution to purine-loading, and no species has a negative PLI for this codon position. Few species achieve these high values in the second and third positions. Also in keeping with the RNY rule, third codon positions are predominantly pyrimidine-loaded (negative PLIs). From the slopes of the plots, third positions appear most responsive to AG-pressure so that, in species with high PLIs, the PLI of the third codon position becomes positive (i.e. the RNY rule no longer holds). However, in bacteria the slope value for the first codon position (1.08) is not significantly different from that of the third codon position (1.15; P=0.082). As in the case of the response to GC-pressure, the response to AG-pressure is least in the second codon position. Nevertheless, AG-pressure is sufficient to affect the amino acid content of encoded proteins (i.e. species with high PLIs have more amino acids encoded by purine-rich codons; Lao and Forsdyke 2000). In general, phages are less responsive to AG-pressure than bacteria in first codon positions (slope of 0.95 versus 1.08; P = 0.04), and more responsive in third codon positions (slope of 1.32 versus 1.15; P = 0.02). Second codon positions show no significant difference (0.73 versus 0.77; P = 0.58). We have noted above that as the PLI increases, an increase in the T bases diminishes the contribution of the W bases to the PLI in bacteria (Fig. 2b), but not in phage (Fig. 2a). For each of the three codon position there are four possible bases. Similar plots to those shown in Figs. 3c,d were constructed for each base at each codon position (e.g. T1, T2, T3). The linear regression parameters are shown in Table. 1. Differences in T bases are largely due to position T3 detracting from responses to AG-pressure in bacteria, but not in phage (slopes of 1.3 and -0.2, respectively). Similarly, as the PLI increases, a decrease in the G bases diminishes the contribution of the S bases to the PLI in bacteria (Fig. 2d), but this diminution is barely significant in phage (Fig. 2c). This is largely due to position G3 detracting from responses to AG-pressure in bacteria more than in phage (slopes of -1.12 and -0.23, respectively). While all codon positions contribute significantly to the differences in slopes between phage and bacteria, such differences are least in the case of second codon position pyrimidines (T2, C2). |
|
Table 1. Parameters of linear
regressions for contributions of bases in |
||||||||||||||||||
|
Organ- |
Codon |
W-bases |
S-bases |
|||||||||||||||
|
A |
T |
G |
C |
|||||||||||||||
|
1a |
2 |
3 |
1 |
2 |
3 |
1 |
2 |
3 |
1 |
2 |
3 |
|||||||
|
|
||||||||||||||||||
|
Phage
|
|
Y0b |
|
272.2 |
304.5 |
199.8 |
|
197.9 |
279.5 |
340.2 |
|
327.5 |
174.3 |
221.4 |
|
202.1 |
241.7 |
238.6 |
|
|
Slopec |
|
0.46 |
0.46 |
0.89 |
|
-0.15 |
-0.06 |
-0.2 |
|
0.02 |
-0.1 |
-0.23 |
|
-0.32 |
-0.31 |
-0.45 |
|
|
P |
0.021 |
0.06 |
0.67 |
0.0014 |
0.009 |
|||||||||||||
|
|
r2 |
|
0.41 |
0.5 |
0.47 |
|
0.1 |
0.03 |
0.016 |
|
<0.0001 |
0.06 |
0.04 |
|
0.25 |
0.43 |
0.12 |
|
|
Bact-
|
|
Y0 |
|
226.3 |
269.4 |
122.4 |
|
151.9 |
281.9 |
174.6 |
|
375.6 |
193.4 |
312.9 |
|
246.2 |
255.3 |
390 |
|
Slope |
0.84 |
0.72 |
1.7 |
0.23 |
0.037 |
1.3 |
-0.3 |
-0.33 |
-1.12 |
-0.77 |
-0.42 |
-1.87 |
||||||
|
|
r2 |
|
0.61 |
0.65 |
0.62 |
|
0.14 |
0.005 |
0.36 |
|
0.15 |
0.4 |
0.4 |
|
0.6 |
0.43 |
0.54 |
|
|
|
||||||||||||||||||
|
|
|
P valued |
|
<0.0001 |
<0.0001 |
<0.0001 |
|
<0.0001 |
0.006 |
<0.0001 |
|
<0.0001 |
<0.0001 |
<0.0001 |
|
<0.0001 |
0.002 |
<0.0001 |
|
a Codon positions 1, 2 or 3. |
||||||||||||||||||
|
b Y0, intercept on Y-axis when value of X-axis is zero. r2, adjusted coefficient of determination. |
||||||||||||||||||
|
c Probabilities that slope values are not different from zero are all <0.0001, except as shown. |
||||||||||||||||||
|
d Probabilities that phage slope values are not significantly different from the corresponding bacterial slope values. |
||||||||||||||||||
|
Inverse relationship between PLI and (G+C)% The response of each individual base to GC-pressure is shown in Fig. 4. At low values of (G+C)%, A is greater than T, and G is greater than C (i.e. there is purine-loading). As (G+C)% increases, so G and C increase, and A and T decrease. However, C increases more than G, and A decreases more than T, so that whereas differences between the members of Chargaff base pairs are high at low (G+C)%, these differences (ΔW and ΔS) become less as (G+C)% increases (i.e. the regression plots converge). Thus, Chargaff differences tend to zero at high values of (G+C)%, and there is an inverse relationship between (G+C)% and the PLI. |
 |
| Fig. 4. Relative responses of different bases to GC-pressure in phage (A) and bacteria (B). Symbols are as in Fig. 2. Note that the sum of Y0 values is zero for the S bases and 1000 for the W bases. |
This was further analyzed in terms of the contributions of bases in individual codon positions. Plots similar to those of Fig. 4 were prepared for each base (i.e. instead of plotting all A bases, A1, A2 and A3 were plotted separately; Table 2). As indicated by the slopes of the regression lines (negative for the W bases and positive for the S bases), in all cases the bases in the third codon positions are most responsive to (G+C)% pressure (the pressure to increase the S bases at the expense of the W bases). Phages tend to be more responsive than bacteria in second codon positions, and less responsive than bacteria in the case of third codon position pyrimidines (T3, C3). |
|
Table 2.
Parameters of linear regression plots showing contributions |
||||||||||||||||||
| . |
W bases |
S bases |
||||||||||||||||
|
A |
T |
G |
C |
|||||||||||||||
|
Organ |
Codon position |
1 |
2 |
3 |
1 |
2 |
3 |
1 |
2 |
3 |
1 |
2 |
3 |
|||||
|
|
||||||||||||||||||
|
Phage |
|
Y0 |
|
514 |
507 |
650 |
|
297 |
332 |
700 |
|
203 |
60 |
-148 |
|
-13 |
100 |
-202 |
|
Slope |
-4.82 |
-3.92 |
-8.92 |
-2.5 |
-1.28 |
-8.56 |
2.88 |
2.45 |
8.1 |
4.43 |
2.75 |
9.38 |
||||||
|
|
|
r2 |
|
0.76 |
0.59 |
0.78 |
|
0.45 |
0.27 |
0.64 |
|
0.51 |
0.65 |
0.89 |
|
0.8 |
0.56 |
0.87 |
|
Bact- |
|
Y0 |
|
501 |
480 |
687 |
|
282 |
329 |
721 |
|
217 |
71 |
-131 |
|
0.15 |
120 |
-277 |
|
Slope |
-4.57 |
-3.43 |
-9.41 |
-2.3 |
-0.87 |
-9.4 |
2.8 |
2.06 |
7.58 |
4.07 |
2.25 |
11.2 |
||||||
|
|
|
r2 |
|
0.86 |
0.72 |
0.93 |
|
0.68 |
0.17 |
0.92 |
|
0.62 |
0.73 |
0.9 |
|
0.82 |
0.6 |
0.94 |
|
|
||||||||||||||||||
|
|
|
P valuea |
|
0.26 |
0.06 |
0.12 |
|
0.32 |
0.07 |
0.02 |
|
0.74 |
0.008 |
0.08 |
|
0.11 |
0.02 |
<0.0001 |
|
a Probability that slope value for phage is not different from slope value for bacteria. |
||||||||||||||||||
The conflict between AG-pressure and GC-pressure is further shown in Figs. 5 a,b where PLI values are plotted against (G+C)% values. As in some other taxa (Lao and Forsdyke 2000; Cristillo et al 2001), as (G+C)% increases, the PLI decreases. Regression lines cross the abscissa at 68% G+C (phage) and 65.5% G+C (bacteria), so that at high (G+C) percentages PLIs tend to become negative (i.e. pyrimidine-loading). Whereas for bacteria this pyrimidine-loading appears as a direct correlate of GC-pressure, appearing mainly at high values of (G+C)% (r2 = 0.53), certain bacteriophage species with intermediate (G+C)% values have high pyrimidine-loading (r2 = 0.13; Fig. 5a). Each of the latter species is represented by several ORFs, so this is unlikely to be due to the selective deposition in GenBank of non-representative ORFs that happen, by chance, to have these features. |
 |
| Fig. 5. Conflict between GC-pressure and AG-pressure in phage (A, C)
and bacteria (B, D). AG-pressure is expressed either as the combined Chargaff differences (PLI; A, B), or as the separate Chargaff differences (ΔW and ΔS; C, D). In A and B, PLI values are zero when (G+C)% values are 68.2 and 65.5, respectively. In C and D, W base Chargaff differences (open circles) are zero when (G+C)% values are 64.1 and 69.7, respectively; S base Chargaff differences (filled squares) are zero when (G+C)% values are 73.8 and 61.5, respectively. Note that the sums of Y0 values in C and D, equal the Y0 values of A and B, respectively. |
| Chargaff differences both for the W bases and for the S bases are affected by GC-pressure (Figs. 5c, d). The slopes of the plots of ΔW and ΔS against (G+C)% are not significantly different from each other both in the case of phages (-1.78 versus -1.04, respectively; P = 0.154), and bacteria (-1.61 versus -1.71, respectively; P = 0.48). However, whereas the slope for ΔW is not different between bacteriophage and bacteria (P = 0.62), the slope for ΔS is less for bacteriophage than for bacteria (P = 0.003). |
 |
| Fig. 6. Influence of GC-pressure on purine-loading at different codon positions. PLI values for each codon position are plotted against (G+C) percentages of the same codon position. Symbols are as in Fig. 3. |
|
The negative effect of GC-pressure on PLI values varies
with the codon position. In Fig. 6 PLI values for each codon position are
plotted against the corresponding (G+C)% values for the
same
codon
position. Although codon position 2 is the position least responsive to either
GC- or AG-pressures (Fig. 3),
it is the position most responsive to the effects of GC-pressure on purine-loading
(i.e. linear regression plots for position 2 have the greatest negative slope).
First codon positions of phage and bacteria acquire pyrimidines as (G+C)%
increases, but sustain their overall net purine-loading in accord with the RNY
rule.
On the other hand, codon position 3, the position most responsive to GC- and AG-pressures (Fig. 3), is the position least responsive to the effects of GC-pressure on purine-loading (i.e. linear regression plots for position 3 have the least negative slopes). Already predominantly pyrimidine-loaded in accord with the RNY rule, phage third positions, on average, do not acquire significantly more pyrimidines as (G+C)% increases (slope = -0.47; P = 0.36). However, bacterial third positions show some increase in pyrimidine-loading as (G+C)% increases (slope = -1.93; P < 0.0001). This third codon position difference between phage and bacteria is significant (P = 0.0004), whereas differences with respect to first and second positions are not significant (P > 0.05). |
 |
| Fig. 7. Responses to AG-pressure of W and S base Chargaff differences for
all codon positions (A, B) and for individual codon positions (C-F).
Data for phage (A, C, E). Data for bacteria (B, D, F). In A and B open circles refer to W base Chargaff differences and filled squares refer to S base Chargaff differences. Symbols for C-F are as in Fig. 3. Note that, in each of A and B, Y0 values sum to zero. Similar, the sum of Y0 values in C (-41) cancels out the sum of Y0 values in E (+41), and the sum of Y0 values in D (+9.6) cancels out the sum of Y0 values in F (-9.6). |
|
W and S base contributions to PLIs W base Chargaff differences appear most responsive to AG-pressure. The slopes of regression plots for ΔW against PLI are greater than those for ΔS against PLI (P < 0.0001 for both phage and bacteria). For phage (Fig. 7a), the regression lines intersect at a PLI value of 56.7 bases/kb, so that, for phage species with low and negative PLI values, ΔS makes the greatest contribution. For bacteria (Fig. 7b) the regression lines intersect at a PLI value of -49.2, so ΔW makes the greatest contribution for most bacterial species. As shown above (Figs. 3c,d), different codon positions make different contributions to the PLI. This was broken down into its ΔW and ΔS components (Figs. 7c-f). In phages (Figs. 7c,e) first codon position ΔW values are twice as responsive to AG-pressure as the ΔS values (slopes of 0.61 and 0.34, respectively). This difference is less in bacteria (Figs. 7d,f; slopes of 0.61 and 0.47, respectively). While the slopes for first codon position ΔW values for phage and bacteria are identical (0.61), slopes for first position ΔS values (0.34 and 0.47) differ significantly (P = 0.013). In both phage and bacterial species, second codon positions tend to be C-rich (negative ΔS values), and to respond least to AG-pressure (slopes of 0.21 for phage, and 0.08 for bacteria, which are significantly different; P = 0.0006). In phages third position ΔW values are most responsive to AG-pressure (slope 1.09 versus 0.40 in bacteria), whereas in bacteria third position ΔS values are most responsive (slope 0.75 versus 0.23 in phage). |
 |
|
Fig. 8. Responses to AG-pressure of W and S base Chargaff differences for
different codon positions. Quadrant plots for phage (A) and bacteria (B).
AG-pressure increases from bottom left to top right. The regression line for all codon positions extends to both axes, but symbols are not shown. Regression lines for individual codon positions do not extend to both axes, and symbols are shown as in Fig. 3. |
|
The relative responses of ΔW and ΔS values to AG pressure can also be displayed in "quadrant" plots where, for each species, the ΔW value is plotted against the ΔS value (Fig. 8). Here the path towards maximum purine-loading traces a diagonal from the lower left quadrant (TC-richness), where most of the points for third codon positions lie, to the upper right quadrant (AG-richness), where most of the points for first codon positions lie. Most points for second codon positions lie in the upper left quadrant (AC-richness). Thus, the RNY rule is supported, with N (any base) actually being either A or C (i.e. an "RMY rule" where M represents A or C). For all codon positions, the overall slope for phage is twice that for bacteria (0.85 and 0.42, respectively; P = 0.0006). Thus, for every ΔS unit added to their coding sequences, phages add twice as many ΔW units as bacteria. Second codon positions in the AC-quadrant make a major contribution to this difference (phage slope value of 0.47 versus bacterial slope value of -0.03; P = 0.003). Here, for phage (Fig. 8a), enrichment for A relative to T is accompanied by a tendency to loose C and gain G. However, for bacteria (Fig. 8b), the tendency to loose C and gain G is not accompanied by enrichment for A relative to T, and the slope of the linear regression plot is not significantly different from zero (P = 0.55). Thus, on average, in bacteria second codon position ΔS values respond to AG-pressure, but ΔW values do not. In the case of third codon positions, a lack of positive responsiveness of ΔW values to AG- pressure is found both for phage and bacteria. The slope for phage (-0.01) is not significantly different from zero (P = 0.94), whereas the slope for bacteria, although slight (-0.08) is significantly negative (P = 0.0002), indicating that as G-loading greatly increases, A-loading decreases slightly. Linear regression plots for the AG-rich first codon positions do not have a significantly greater slope for phage than for bacteria (P = 0.10).
Pressures exerted by collectivities of bases, (G+C) and (A+G), are gaining recognition as playing important functional roles (Forsdyke and Mortimer 2000; Pizzi and Frontali 2001; Forsdyke, 2002a). Yet, as seen above, the individual members of each Chargaff pair, not generally recognized as exerting individual pressures, have effects that are often not proportionate to those of their Chargaff partners. Thus, the individual bases can be considered as exerting unique pressures. A-pressure, C-pressure, G-pressure and T-pressure can be considered as distinct entities. As in the cases of GC-pressure and AG-pressure, we investigated both the contribution of each codon position to each base pressure, and the response of each codon position to that pressure (slopes of regression plots). |
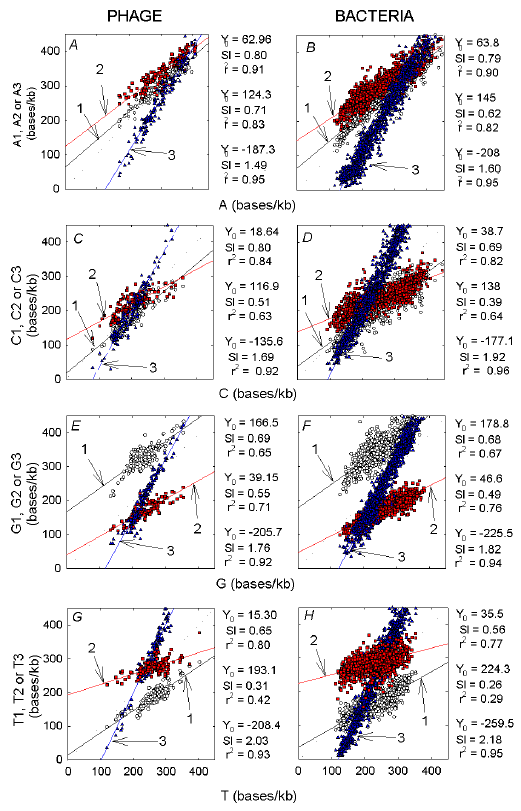 |
|
Fig. 9. Contributions of individual bases in different codon positions to
overall pressures exerted by the corresponding bases in phage (A, C,
E, G) and bacteria (B, D, F, H).
Adenine, A, B; cytosine, C, D; guanine, E, F; thymine G, H. Symbols for different codon positions are as in Fig. 3. Parameters of the linear regression lines for first, second and third codon positions (in descending order) are to the right of each graph. |
|
In general, third codon positions respond most to the corresponding base pressure, and second codon positions respond least (Fig. 9; see also Besemer and Borodovsky 1999). Whereas A-pressure is contributed mostly by the second codon position in low A species, the contributions of the three codon positions become equal in high A species (i.e. the linear regression plots converge; Figs. 9a,b). The plots for C pressure have similar characteristics to those for A pressure, except that the convergence occurs at low C levels in the third codon position, so that this position makes the major contribution to C pressure in species with high C levels (Figs. 9c,d). G in the first codon position generally makes a greater contribution to R pressure than does A in this position, so the RNY rule tends to be a "GNY rule" (Figs. 9e,f; see also Kypr and Mrazek 1987). Only in species under high G-pressure does the contribution of the third codon position approach that of the first codon position. In all species the first codon position contributes few Ts when T-pressure is low, but it responds better than the second codon position to T-pressure (slope of 0.65 versus 0.31; Figs. 9g,h). In general, phages respond better than bacteria to the pressures of the individual bases in the cases of the first and second codon positions (slopes of phage plots greater or equal to that of bacteria plots). The converse applies for the third codon positions.
The above description of the general characteristics of the purine-loading phenomenon for phage and bacteria provides a frame of reference for assessing the role of purine-loading in cases of individual phages and their specific hosts. To begin this, 28 bacterial species hosting 48 bacteriophage species were identified with minimal ambiguity from GenBank records. Bacteriophages and their hosts were found to correspond closely in (G+C) percentages (r2 = 0.90; Marmur and Greenspan 1963), but much less so in purine-loading (r2 = 0.27). For example, the (G+C)% of Corynephage beta (an average of 47.3 for the 6 ORFs available in GenBank) is quite close to that of its host Corynebacterium diphtheriae (an average of 53.2 for the 27 available ORFs), whereas its PLI (105.2) differs dramatically, the host being pyrimidine-loaded (PLI of -8.5). Whereas Vibrio cholerae (4405 ORFs in GenBank) has a PLI of 28.3, two of its phage (fs-1 with 15 available ORFs; fs-2 with 9 available ORFs) are pyrimidine-loaded (PLIs of -62.5 and -45.9 respectively). However, despite these disparities there is a positive correlation between the PLIs of phage and their hosts (P < 0.0001). |
|
As shown for phage lambda (Fig. 1), individual ORFs can vary widely in their degree of purine-loading. A selection of four ORFs from the phage lambda genome's 91 ORFs might not be representative. However, for many purposes it is better to have limited sequence information from a large number of genomes, than to have full sequence information from a limited number of genomes (Forsdyke 2002b). The statistically distinct trends that emerge from the present study of a large number of genomes support our choice of an arbitrary four ORF cut-off point. Because phages and bacteria differ in many of the proteins they encode, this could influence their respective base compositions. However, many of the differences reported here relate to the third positions of codons, where the protein-encoding role ("protein pressure") is not so critical. The proteins of phages and their hosts are synthesized using the same ribosomes and many similar tRNAs (Kunisawa 2000), so that "translational-pressures" (Sharp et al 1993; Gursky and Beabealashvilli 1994) might be similar. One manifestation of translation pressure is the "RNY rule", which could relate to the fact that a major component of the interaction energy between two complementary bases derives from the stacking interactions with their neighbours. Codon-anticodon interactions are more than triplet-triplet interactions. By involving the neighbouring bases they tend to be quintuplet-quintuplet interactions. Triplet anticodon sequences in tRNAs vary according to the amino acid specificity of the tRNA, but there are regularities in the flanking bases that appear to play a role in tRNA-mRNA interactions (Bossi and Roth 1980; Simonson and Lake 2002). On one side of tRNA anticodons there is a relatively invariant pyrimidine and on the other side there is a relatively invariant purine. It would seem advantageous for a codon in mRNA when interacting with its cognate anticodon to be preceded in the mRNA by an NNY codon (so that the Y can pair with the invariant purine), and to be followed in the mRNA by an RNN codon (so that the R can pair with the invariant pyrimidine). Since, codons are translated successively, it follows that codons which, while encoding amino acids correctly, can also following the RNY rule, will be preferred (Eigen and Schuster 1978). There is a potential conflict between pressure to encode certain amino acids (protein pressure) and this "RNY-pressure." These pressures also have to accommodate to GC- and AG-pressures. Indeed, in species under high AG-pressures third codon positions tend to acquire purines and the RNY rule no longer applies (Figs. 3c,d). Purine-loading as a viral adaptation Comparisons of the genomes of various eukaryotic viruses and their hosts have shown that some viruses differ dramatically from their hosts in (G+C)%. Furthermore, some purine-load their RNAs to exceed greatly that of their hosts (e.g. the retrovirus HIV-1). Others pyrimidine-load their RNAs (e.g. HTLV-1, a retrovirus with a profounder degree of latency than HIV-1). Another latent virus (Epstein-Barr) pyrimidine-loads the majority of its RNAs, which are not expressed in the latent phase, but purine-loads the main RNA expressed in all forms of latency. It is argued that each virus has a strategy related to its host, part of which is reflected in differences in base composition (Bell and Forsdyke 1999b; Cristillo et al 2001). If true for eukaryotic viruses, similar considerations might apply to prokaryotic viruses. To approach this systematically, we have first sought general distinctions between phage and bacterial genomes. Such distinctions are observed, both regarding single bases (Figs. 2, 4), and codon positions (Figs. 3, 6-9; Tables 1, 2). Although the number of phage and bacteria species represented currently in GenBank are but a small sub-set of the total set of such species on the planet, they are derived from diverse environments and so may prove to be fairly representative. Many of the differences are apparent at the third codon position, suggesting that the basis of the difference does not lie in the need to synthesize different proteins, but relates to base composition per se. However, there are differences in first and second codon positions, which are sufficient to affect the amino acid compositions of proteins. Thus, there are circumstances under which it is of greater advantage to have a particular base in a particular position in a nucleic acid, than to have a particular amino acid in a particular position in a protein. The pressure to encode a particular amino acid is great, but it can be over-ridden by other pressures (Sueoka 1961; Rocha et al 1999; Mackiewicz et al 1999; Tillier and Collins 2000; Lao and Forsdyke 2000). Inverse relationship between purine-loading and (G+C)% There is a conflict between AG-pressure and GC-pressure (Figs. 4-6). There are two ways to modulate (G+C)% when the total number of bases is constant; either by changing the number of G's, or by changing the number of C's. As (G+C)% increases, trading A for G would not affect the PLI. Likewise, trading T for C would not affect the PLI. However, if T were replaced by G, the PLI would increase as (G+C)% increases. Conversely, if A were replaced with C the PLI would decrease as (G+C)% increases. The latter inverse relationship is observed in certain bacteria (Lao and Forsdyke 2000), and certain eukaryotes (Cristillo et al 2001; Saccone et al 2000), and is confirmed here. The negative slope (Figs. 5a,b) indicates a preference for trading A with C in order to modulate GC content; this contributes to an inverse relationship between (G+C)% and ΔW and between (G+C)% and ΔS (Figures 5c,d). GC-pressure appears as the superior pressure in that AG-pressure is eliminated at high values of (G+C)%. Being already highly Y-loaded, third codon positions are less in a position to give up A and acquire C, and so appear less responsive to this aspect of GC-pressure than the first and second codon positions (Figs. 6a,b). Being already highly R-loaded, first codon positions should be best able to trade A for C, but second codon positions are more responsive than first positions in this respect (slopes of -6.72 versus -4.31 in the case of phage, and -6.38 versus -4.70 in the case of bacteria). Low values for (G+C)% when the PLI is high (Fig. 5) suggest the superior power of AG-pressure in these circumstances. Indeed, we have postulated that the PLI should increase with optimum growth temperature, and have shown this to occur in bacteria and to be accompanied by a small, but significant decrease in (G+C)% (Lao and Forsdyke 2000; Lambros et al. 2003). Hence, a causal chain of evolutionary events might be (i) need to adapt to high temperature: -->(ii) purine-loading: -->(iii) decrease in (G+C)%. We have speculated elsewhere on the roles purine-loading plays in the economy of a cell and its pathogens (Cristillo et al., 2001; Forsdyke et al 2002). Whatever that role, it seems unlikely that A and G would function equally in this respect. The magnitude of the function would depend on the relative contributions of the W bases and of the S bases to the PLI. A better measure might be to express the PLI in units of ΔA (i.e. PLIA). Thus, PLIA = ΔA + M (ΔG), where M is a constant, and ΔA = - M (ΔG) + PLIA. In the latter case, a plot of ΔA against ΔG should provide values for -M and PLIA. We have found that such plots (Fig. 8) do not provide constant values for M, but M is usually less than one, indicating that the S bases are less effective than the W bases in contributing to whatever function(s) is served by purine-loading. Currently we conceive of this as a repelling function (the avoidance of "kissing" interactions; Bell and Forsdyke 1999b), so ΔW and ΔS values might reflect the ability of bases not to engage in base interactions other than the classical Watson-Crick interactions. Consistent with a greater role in this respect, the W bases appear most responsive to AG-pressure, and make the greatest contribution in most bacterial species (Fig. 7b). However, in phage species with low PLI values, the S bases make the greatest contribution (Fig. 7a). Remarkably, ΔW3 is most responsive to AG-pressure in phage (i.e. increase in A and/or decrease in T; Fig. 7c), whereas ΔS3 is most responsive in bacteria (i.e. increase in G and/or decrease in C; Fig. 7f). The disjoint nature of the ΔW and ΔS responses is evident in quadrant plots, where both in phage and bacteria ΔW3 hardly changes as ΔS3 responds to AG-pressure (Fig. 8). Our results have prepared us for analysis of relationships between subsets of phages (e.g. temperate versus lysogenic) and their hosts, and between individual phages and their hosts (Blaisdell et al 1996). For example, although phage and bacteria, in general, responding differentially to GC-pressure at different codon positions (Table 2), there is a general correlation (r2 = 0.90) between the (G+C)% of a phage and that of its host, which is much better than the correlation between PLIs (r2 = 0.27). Since third codon positions are the most responsive to AG-pressure, it seems likely that, like GC-pressure, AG-pressure functions at the nucleic acid level, rather than at the level of the encoded protein. Evidence is presented elsewhere (Forsdyke, 1998) that very small differences between DNAs in (G+C)% are sufficient to impair the function to which (G+C)% is likely to relate (recombination between those DNAs; Forsdyke and Mortimer 2000). Thus it is possible that, although (G+C)% values for phages and their hosts are generally correlated, the observed differences (albeit small) are still sufficient for this role. While there may be disagreement about the causes of regularities in base composition (Forsdyke and Mortimer 2000), the existence of such regularities (e.g. the results of "GC-pressure" or "AG-pressure") is not in question. AG-pressure is now receiving attention as an indicator of transcription direction (Saul and Battistuta 1988; Dang et al 1998; Bell and Forsdyke 1999b), and as an explanation for low-complexity sequences in proteins of P. falciparum (Pizzi and Frontali 2001; Forsdyke 2002a), and herpes viruses (Cristillo et al 2001). Using the graphical approach applied by Muto and Osawa (1987) to the analysis of GC-pressure, we have analyzed pressures on DNA sequences with particular emphasis on AG-pressure. We have confirmed with a large data set the previously observed reciprocal relationship between GC-pressure and AG-pressure (Lao and Forsdyke 2000; Forsdyke and Mortimer 2000; Saccone et al 2000), and have shown this to follow from the preferential trading of certain bases. We have observed differential responses of bacteriophages and bacteria to pressures on base compositions. There might be similar differences between eukaryotic cells and their viruses that could be exploited therapeutically. |
|
Sequences Programs for Microsoft Excel were written in Microsoft Visual Basic to calculate base compositions from species codon usage tables. The September 2000 release of GenBank had been used to generate "Codon Usage Tables from GenBank" (CUTG) from the pooled ORF sequences of a species (Nakamura et al 2000). We excluded organisms for which there were not available sequences of more than 3 ORFs and 2500 bases. The CUTG database had not been screened for redundancies; thus, while the sequence of Escherichia coli K12 contributed approximately 4300 ORFs, under this heading there are 14780 ORFs in the database, implying at least three independent genome equivalents had been used to derived the 64 codon usage values for the species. While this might have introduced some bias, each of the 1046 bacterial species studied is represented by only one data point in our plots. Thus E. coli with 14780 ORFs has the same representation as the incompletely sequenced genome of Rickettsia typhi (6 ORFs). The CUTG database includes under the heading "bacteria" both archaebacteria and eubacteria. Similarly, the heading "phage" includes both viruses that infect archaebacteria and viruses that infect eubacteria. Chargaff difference analysis Chargaff differences (ΔW, ΔS) are the differences between the numbers of the classical Watson-Crick pairing bases in a nucleic acid segment ("AT-skew", "GC-skew;" Frank and Lobry 1999). The sign of the differences depends on the direction of subtraction, which in some previous work was determined alphabetically, but in the present work is determined by subtracting the number of pyrimidines (Y) from the number of purines (R). Thus, purine excesses (R>Y) score positively and pyrimidine excesses (R<Y) score negatively. Chargaff differences (absolute or %) may be calculated as A-T [or as (A-T)/W], and as G-C [or as (G-C)/S]. Here, A, T, G and C can be the frequency of each base in 1 kb sequence windows, and W = A + T, and S = G + C. This approach makes no assumption about the disposition of open reading frames (ORFs), and can be applied to uncharted DNA. For the importance of 1 kb window sizes and other details see Dang et al (1998) and Bell and Forsdyke (1999a). If the ORFs in a sequence are known, ΔW and ΔS in bases/kb may be calculated either directly from ORF base compositions, or, indirectly, from codon-usage tables. Then ΔW = 1000[(A-T)/N], and ΔS = 1000[(G-C)/N], where N is the total number of bases in an ORF. These two values can be summed to obtain a value for the purine-loading index (i.e. ΔW + ΔS = PLI). This approach disregards non-ORF DNA. To distinguish bases in different codon positions, base letters are followed by codon positions. For example, whereas T refers to the quantity of bases in all three codon positions, T1, T2, and T3 refer to the quantities of T in first, second and third codon positions, respectively. Accordingly, the contribution of first codon positions to the W-base component of the Chargaff difference, ΔW1, would be 1000[(A1-T1)/N1]. For AT-rich genomes, purine-loading is mostly with respect to A, whereas for GC-rich genomes, purine-loading is mostly with respect to G. In organisms with high PLI values (e.g. thermophilic bacteria), purine-loading involves both purines (Lao and Forsdyke 2000). An organism in which one or both Chargaff differences reflect the purine-loading of a significant excess of mRNAs is held to comply with "Szybalski's transcription direction rule" (Bell and Forsdyke 1999b). It should be noted that GC-pressure is assessed as the sum of G + C in a sequence, whereas AG-pressure is assessed as the excess of the R bases over the Y bases. Although it might be preferable to assess GC-pressure and AG-pressure in the same way (bases/kb), we here retain the classical measure of GC-pressure, (G+C)% (i.e. bases/0.1 kb). Base "pressure" must be understood in context. For example, as (G+C)% increases, (A+T)% decreases, so (G+C)% suffices to quantitate both the entity "GC-pressure" and, negatively, the entity "AT-pressure." Similarly, (A+G)% suffices to quantitate both "AG-pressure" and, negatively, "CT-pressure." The PLI is positive when purines are greater than 50% and can be seen as a response to "upward AG-pressure" rather than a response to "downward CT-pressure." (G+C)% increasing above 50% can be seen as a response to "upward GC-pressure" rather than as a response to "downward AT-pressure". (G+C)% decreasing below 50% can be seen as a response to "downward GC-pressure" rather than as a response to "upward AT-pressure". However, we here refer to only one member of a reciprocating base pair (i.e. G+C rather than A+T, and A+G rather than C+T), and simply consider pressures due to single bases, or pairs of bases, as increasing from 0% to 100%. Statistics First-order linear regression analyses were performed with the assumption that data points were normally distributed. The probability that the slopes of two regression lines were not significantly different from each other was calculated using an interaction model with dummy qualitative variables as described previously (Forsdyke 1998). Acknowledgements We thank T. St. Denis for assistance, J. Gerlach for computer configuration, and A. Kropinski for reviewing the manuscript. Access to the GCG software suite was provided by the Canadian Bioinformatics Resource, Halifax. The National Research Council of Canada, Academic Press, Cold Spring Harbor Laboratory Press and Elsevier Science gave permission for the inclusion of full-text versions of relevant preceding papers at our internet site. |
|
Bell SJ, Forsdyke DR. 1999a Accounting units in DNA. J. Theor. Biol. 197: 51-61. Bell SJ, Forsdyke DR. 1999b Deviations from Chargaff's second parity rule correlate with direction of transcription. J. Theor. Biol. 197: 63-76. Besemer J, Borodovsky M 1999 Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 27: 3911-3920. Blaisdell BE, Cambell AM, Karlin S. 1996 Similarities and dissimilarities of phage genomes. Proc. Natl. Acad. Sci. USA 93: 5854-5859. Bossi L, Roth JR 1980 The influence of codon context on genetic code translation. Nature 286: 123-127. Brenner S. 1991 Summary and concluding remarks. In Osawa S, Honjo, T, eds. Evolution of Life. Fossils, Molecules and Culture. Berlin: Springer-Verlag, p 453-456. Cristillo AD, Mortimer JR, Barrette IH, Lillicrap TP, Forsdyke DR. 2001 Double-stranded RNA as a not-self alarm signal: to evade, most viruses purine-load their RNAs, but some (HTLV-1, EBV) pyrimidine-load. J. Theor. Biol. 208: 475-491. Dang KD, Dutt PB, Forsdyke DR. 1998 Chargaff differences correlate with transcription direction in the bithorax complex of Drosophila. Biochem. Cell Biol. 76: 129-137. Eigen M, Schuster P. 1978 The hypercycle. A principle of natural self-organization. Part C. The realistic hypercycle. Naturwissenschaften 65: 341-369. Forsdyke DR 1998 An alternative way of thinking about stem-loops in DNA. A case study of the human G0S2 gene. J. Theor. Biol. 192: 489-504. Forsdyke DR 2002a Selective pressures that decrease synonymous mutations in Plasmodium falciparum. Trends Parasitol. 18: 411-418. Forsdyke DR. 2002b Symmetry observations in long nucleotide sequences. Bioinformatics 18: 215-217. Forsdyke DR, Madill CA, Smith SD. 2002 Immunity as a function of the unicellular state: implications of emerging genomic data. Trends Immunol. 23: 575-579. Forsdyke DR, Mortimer JR. 2000 Chargaff's Legacy. Gene 261: 127-137. Frank AC, Lobry JR 1999 Asymmetric substitution patterns: a review of possible underlying mutational or selective mechanisms. Gene 238: 65-77. Gursky YG, Beabealashvilli RSh. 1994 The increase in gene expression induced by introduction of rare codons into the C terminus of the template. Gene 148: 15-21. Hedrix RW, Smith MCM, Burns RN, Ford ME, Hatfull GF. 1999 Evolutionary relationships among diverse bacteriophages and prophages: all the world's a phage. Proc. Natl. Acad. Sci. USA 96: 2192-2197. Knight RD, Freeland SJ, Landweber LF. 2001 A simple model based on mutation and selection explains trends in codon and amino acid usage and GC composition within and across genomes. Genome Biol. 2: research0016.1-0016.13 Kunisawa T. 2000 Functional role of mycobacteriophage transfer RNAs. J. Theor. Biol. 205: 167-170. Kypr J, Mrazek J. 1987 Occurrence of nucleotide triplets in genes and secondary structure of the coded proteins. Int. J. Biol. Macromol. 9: 49-53. Lambros RJ, Mortimer JR, Forsdyke DR. 2003 Optimum growth temperature and the base composition of open reading frames in prokaryotes. Extremophiles (in press) Lao PJ, Forsdyke DR. 2000 Thermophilic bacteria strictly obey Szybalski's transcription direction rule and politely purine-load RNAs with both adenine and guanine. Genome Res. 10: 228-236. Mackiewicz P, Gierlik A, Kowalczuk M, Dudek MR, Cebrat S. 1999 How does replication-associated mutation pressure influence amino acid composition of proteins? Genome Res. 9: 409-416. Marmur J, Greenspan CM. 1963 Transcription in vivo of DNA from bacteriophage SP8. Science 142: 387-389. Muto A, Osawa S. 1987 The guanine and cytosine content of genomic DNA and bacterial evolution. Proc. Natl. Acad. Sci. USA 84: 166-169. Nakamura Y, Gojobori T, Ikemura T. 2000 Codon usage tabulated from the international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 28: 292. Pizzi E, Frontali C. 2001 Low-complexity regions in Plasmodium falciparum proteins. Genome Res. 11: 218-229. Rocha EPC, Danchin A, Viari A. 1999 Universal replication biases in bacteria. Mol. Microbiol. 32: 11-16. Saccone C, Gissi C, Lanave C, Larizza A, Pesole G, Reyes A. 2000 Evolution of the mitochondrial genetic system: an overview. Gene 261: 153-159. Saul A, Battistuta D. 1988 Codon usage in Plasmodium falciparum. Mol. Biochem. Parasitol. 27: 35-42. Schaap T. 1971 Dual information in DNA and the evolution of the genetic code. J. Theor. Biol. 32: 293-298. Schattner P 2002 Searching for RNA genes using base-composition statistics. Nucleic Acids Res. 30: 2076-2082. Sharp PM, Stenico M, Peden JF, Lloyd AT. 1993 Codon usage: mutational bias, translation selection, or both? Biochem. Soc. Trans. 21: 835-841. Shepherd JCW. 1981 Method to determine the reading frame of a protein from the purine/pyrimidine genome sequence and its possible evolutionary justification. Proc. Natl . Acad. Sci. USA. 78: 1596-1600. Simonson AB, Lake JA. 2002 The transorientation hypothesis for codon recognition during protein synthesis. Nature 416: 281-285. Smithies O, Engels WR, Devereux JR, Slightom JL, Shen S. 1981 Base substitutions, length differences and DNA strand asymmetries in the human Gl and Al fetal globin gene region. Cell 26: 345-353. Sueoka N 1961 Compositional correlations between deoxyribonucleic acid and protein. Cold Spring Harb. Symp. Quant. Biol. 26: 35-43. Summers WC. 2001 Bacteriophage therapy. Ann. Rev. Microbiol. 55: 437-452. Szybalski W, Kubinski H, Sheldrick P. 1966 Pyrimidine clusters on the transcribing strands of DNA and their possible role in the initiation of RNA synthesis. Cold Spring Harb. Symp. Quant. Biol. 31: 123-127. Szybalski W, Bovre K, Fiandt M, Guha A, Hradecna Z, Kumar S, Lozeron HA, Maher VM, Nijkamp, HJJ, Summers WC et al 1969 Transcriptional controls in developing bacteriophages. J.Cell Physiol. 74, suppliment 1: 33-70. Tillier ERM, Collins RA. 2000 Replication orientation affects the rate and direction of bacterial gene evolution. J. Mol. Evol. 51: 459-463. Xue HY, Forsdyke DR. 2003 Low-complexity segments in Plasmodium falciparum proteins are primarily nucleic acid level adaptation. Mol. Biochem. Parasitol. 128: 21-32. |
End Note (Feb 2010) Correlations between Phages and Hosts
| A
similar study by Xia & Yuen (2005) noted a
pioneering study by Gibbs & Primrose (1976) that we missed above.
Gibbs A, Primrose S (1976) A correlation between the genome compositions of bacteriophages and their hosts. Intervirology 7, 351-355. Xia X, Yuen K-Y (2005) Differential selection and mutation between dsDNA and ssDNA phages shape the evolution of their genomic AT percentages. BMC Genetics 6:20 |
End Note (Feb 2013) R-Loading due to Selection
|
Many full genome
sequences subsequently become available,
and Charneski et al. (2011) studied the
R-loading (A/T skew) of Staphylococcus aureus,
a member of the Firmicute phylum of
bacteria; but the corresponding bacteriophage predators were excluded.
While heavily influenced by Sueoka's "mutational bias" mode of thought,
they were prepared to acknowledge the influence of some selective "force"
enriching for the purine A, but this did not extend this
to bacterial defence against bacteriophages. They confirm that, following
the RNY codon rule, the first and, less so, second,
codon positions are best able to contribute the extra A
bases. Third bases (and intergenic bases, which we did not study) also
contribute, but weakly. Charneski CA, Honti F, Bryant JM, Hurst LD, Feil EJ (2011) Atypical AT skew in Firmicute genomes results from selection not mutation. PLOS Genetics 7, e1002283. |
![]()
Go to Elementary Principles (2004) Click Here
Go to Bioinformatics Index Click Here
Go to Home Page Click Here
![]()
This page was last edited on 07 Nov 2020 by Donald Forsdyke