|
Optimum growth temperature and the base
composition of open reading frames in prokaryotes
R. J. Lambros, J. R. Mortimer, D. R. Forsdyke (2003)
Extremophiles 7,
443-450. With copyright permission from Springer-Verlag, which provides the original at http://www.springerlink.com
Received by F. T. Robb 30th January 2003. Accepted for
publication by F. T. Robb 2nd May 2003
[This paper was first submitted to Genome
Research in March 2001 and declined. It was successively
declined by Gene, Journal
of Molecular Evolution, Journal of
Theoretical Biology, and Biochemistry
and Cell Biology (Jan. 2003).]

Introduction
Methods
Optimum growth temperatures
Sequences
Chargaff difference analysis
Statistics
Results
Optimum growth temperature, (G +C )%, and PLI
Contributions of individual bases to
deltaW and deltaS
Discussion
PLI is positively related to growth temperature
(G+C)% is negatively related to growth temperature
(G+C)% is negatively related to PLI
Nucleic acid constraints on phenotype
End-Note 2004
End-Note 2005
End-Note January 2007
End-Note_August_2012
End-Note_August_2015

| Abstract
The
purine-loading
index (PLI) is the difference between the numbers of purines (A + G) and
pyrimidines (T + C) per kilobase of single stranded nucleic acid. By
purine-loading their mRNAs organisms may minimize unnecessary RNA-RNA
interactions and prevent inadvertent formation of "self"
double-stranded RNA. Since RNA-RNA interactions have a strong
entropy-driven component, this need to minimize should increase as
temperature increases.
Consistent with this, we report for 550 prokaryotic species that optimum
growth temperature is related to the average PLI of open reading frames.
With increasing temperature prokaryotes tend to acquire base A and lose
base C, while keeping bases T and G relatively constant. Accordingly,
while the PLI increases, the (G+C)% decreases.
The
previously observed positive correlation between (G+C)% and optimum
growth temperature, which applies to RNA species whose structure is of
major importance for their function (ribosomal and transfer RNAs) does
not apply to mRNAs, and hence is unlikely to apply generally to genomic
DNA.
Keywords Base composition
(G+C)%
Growth temperature Purine-loading Thermophiles |

Introduction
|
In most species mRNAs are
purine-loaded in regions with the potential to form
loops in stem-loop secondary structures (Szybalski et al. 1966; Smithies et al.
1981; Bell and Forsdyke 1999a,b). Like GC-pressure and fold (stem-loop)
pressure, the pressure to purine-load (AG-pressure) can affect the amino acid
composition of proteins (Forsdyke and Mortimer
2000). Whereas selective forces
promoting the development of GC and fold pressures are likely to operate
primarily at the genomic level (Grantham 1980; Bernardi and Bernardi 1986;
Forsdyke 1996, 1998, 1999; Barrette et al. 2001; Knight et al. 2001), it is
proposed that selective forces for the development of AG-pressure operate
primarily at the non-genomic level. A general purine-loading of loops in mRNA
secondary structures would militate against unwanted mRNA-mRNA interactions (Bell and Forsdyke
1999b) and the formation of segments of "self"
double-stranded RNA of a length sufficient to trigger intracellular alarms.
Pressure to purine-load provides a possible explanation for apparently
non-functional low complexity (simple sequence) elements in proteins of the
malaria agent Plasmodium falciparum (Pizzi and Frontali 2001; Forsdyke,
2002b; Xue and Forsdyke 2003), and of viruses
(Cristillo et al. 2001).
Being largely entropy-driven
(Lauffer 1975; Cantor and Schimmel 1980),
RNA-RNA interactions should increase with temperature. Thus, the need to prevent
unwanted interactions might be greater in organisms that normally grow at high
temperatures, or are periodically exposed to such temperatures. Consistent with
this, in a previous study of a small number of prokaryotes for which extensive
genomic sequences were available, purine-loading was found to be greatly
increased in thermophiles to an extent sufficient to influence amino acid
composition (Lao and Forsdyke 2000). This was confirmed in a recent study with
25 completely sequenced genomes (Lobry and Chessel
2003). We have now extended
our study to a much larger number of prokaryotic species for which sequence
information, albeit often limited, is available. We have also examined the
responsiveness of bases in different codon positions to AG-pressure and the
relationship of AG-pressure to GC-pressure. Our results have implications for
understanding how nucleic acids maintain active configurations at high
temperatures, and how forces other than those of conventional Darwinian natural
selection can influence protein evolution (Hurst and Merchant 2001; Forsdyke
2001a,b; 2002a,b).
|
Methods
|
Optimum growth temperatures
A table of optimum growth temperatures was obtained from the German
Collection of Microorganisms and Cell Cultures (DSMZ,
Braunschweig). Direct
examination of the primary literature supported the accuracy of the temperatures
in twenty randomly selected cases, so the general accuracy of the table was
assumed.
Sequences
Sequences from the September 2000 release of GenBank have been used to
generate "Codon Usage Tables from GenBank" (CUTG; Nakamura et al.
2000). Programs for Microsoft Excel were written in Microsoft Visual Basic for
Applications to calculate open reading frame (ORF) base compositions from the
CUTG data. We excluded organisms for which there had not been available
sequences of at least four ORFs and 2500 bases. The CUTG database had not been
screened for redundancies; thus, while the sequence of Escherichia coli
K12 contributed approximately 4300 ORFs, under this heading there are 14780 ORFs
in the database, implying at least three independent genome equivalents had been
used to derived the 64 codon usage values for the species. While this might have
introduced some bias, each of the prokaryotic species studied is represented by
only one data point in our plots. Thus E. coli with 14780 ORFs has the
same representation as a genome for which there are only four ORFs in GenBank.
There are reasons why certain species are chosen for sequencing and why certain
ORFs are sequenced first; these may not be representative of the corresponding
genome. Furthermore, some species are closely related phylogenetically, whereas
others are more distantly related. However, the large number of species studied
makes it likely that biases would cancel out to permit a fair general picture.
Indeed, a recent comparison of datasets from completely sequenced and partially
sequenced bacterial genomes gave similar results (Lobry and Chessel
2003).
Chargaff difference analysis
Violating Chargaff's second parity rule
(Forsdyke and Mortimer 2000;
Forsdyke 2002a), Chargaff differences (ΔW,
ΔS) are the differences between the numbers of the classical Watson-Crick
pairing bases in a single-stranded nucleic acid segment
("AT-skew", "GC-skew"). The sign of the differences depends
on the direction of subtraction, which in some previous work was determined
alphabetically, but in the present work is determined by subtracting the number
of pyrimidines (Y) from the number of purines (R). Thus, purine excesses
(R>Y) score positively and pyrimidine excesses (R<Y) score negatively.
Chargaff differences may be calculated as A-T, and as G-C, where A, T, G and
C can be the frequency of each base in 1 kb sequence windows. This approach
makes no assumption about the disposition of ORFs, and can be applied to
uncharted DNA. When an unknown ORF is located, values for windows whose centres
overlap the ORF can be averaged to obtain an approximate value for that ORF. For
the importance of 1 kb window sizes and other details see Bell and Forsdyke
(1999a).
If the ORFs in a sequence are known,
ΔW and
ΔS in bases/kb may be calculated either directly from ORF base
compositions, or, indirectly, from codon-usage tables. Then ΔW =
1000[(A-T)/N], and ΔS = 1000[(G-C)/N], where N is the total number of bases
in an ORF. These two values can be summed to obtain a value for the purine-loading index (i.e. ΔW + ΔS =
PLI). This approach disregards
non-coding DNA. To distinguish bases in different codon positions, base letters
are followed by codon positions. For example, whereas T refers to the
quantity of bases in all three codon positions, T1, T2, and T3 refer to the
quantities of T in first, second and third codon positions, respectively.
Accordingly, the contribution of first codon positions to the W-base component
of the Chargaff difference, ΔW1, would be
1000[(A1-T1)/N1].
It should be noted that
(G+C)% (a measure of "GC-pressure") is
assessed as the sum of G + C in a sequence, whereas Chargaff differences
(a measure of "purine-loading pressure" or "AG pressure")
are assessed as the excess of the R bases over the Y bases. Although it
might be preferable to assess GC-pressure and AG-pressure in the same way
(bases/kb), we here retain the classical measure of GC-pressure (G+C)% (i.e.
bases/0.1 kb).
Statistics
First-order linear regression analyses were performed with the assumption
that data points were normally distributed. The probability that the slopes of
two regression lines were not significantly different from each other was
calculated using an interaction model with dummy qualitative variables as
described previously
(Forsdyke 1998).
|
Results
|
Optimum growth temperature, (G +C )%, and PLI
We found 550 prokaryotic species for which optimum growth temperatures were
known and for which there were at least 4 ORFs and 2500 bases in GenBank. The
average number of ORFs was 144+ or -34. 92 species had less than 6 ORFs. 190 species
had less than 10 ORFs. Optimum growth temperatures ranged from 17oC (Renibacterium
salmoninarum) to 105oC (Pyrodictium occultum). Of the 550 species,
494 had temperature optima below 60oC, and 56 had temperature optima of 60oC
and higher.
|
|
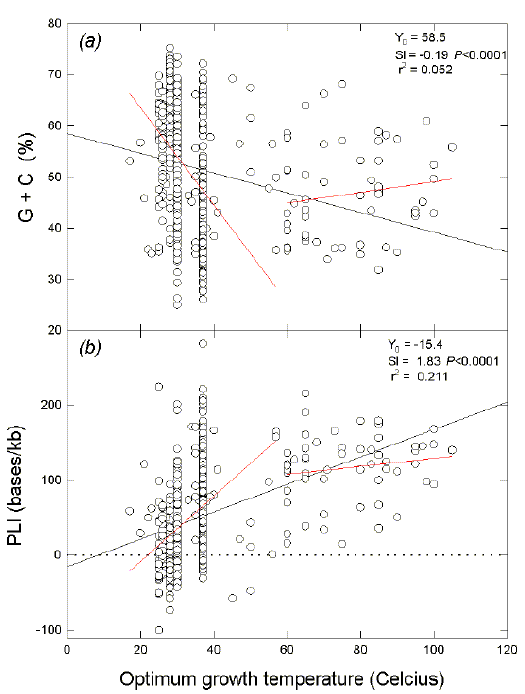
|
|
Fig. 1. Variation of (a) ORF
G+C, and (b) ORF PLI with the temperature
required for optimal growth. The least squares regression lines for the 550
prokaryotic species studied extend to the Y-axis. Y0, value for
intercept at the Y-axis; Sl, slope with associated probability (P) that
the slope is not significantly different from zero; r2, adjusted
square of the correlation coefficient (i.e. the coefficient of determination).
Regression lines for the 494 species with temperature optima <60oC, and for
the 56 species with temperature optima of 60oC and greater, are shown as short
lines, which do not extend to the Y-axis. Data from this figure are included in Table 1.
|
Although the wide scatter of data points indicates other variables affecting
base composition, linear regression plots show that the optimum growth
temperature of prokaryotes is inversely related to ORF (G+C) percentages (Fig.
1a), and directly related to ORF PLIs (Fig. 1b). In both cases, the greatest
response to growth temperature occurs much below 60oC, whereas there is no
significant further response over the higher temperature range (i.e. over the
higher temperature range P values for local slopes were 0.26 and 0.23 for
Figs. 1a and 1b, respectively). The average (G+C)% for prokaryotes with optima
below 60oC (52.0 + or - 0.6), was greater (P = 0.002; unpaired t-test) than
that for prokaryotes with optima of 60oC and higher (46.6 + or - 1.2). The average
PLI for prokaryotes for optima below 60oC (44.4+ or -2.5) was less (P < 0.00001)
than that for prokaryotes with optima of 60oC and higher (116.3 + or - 6.0). Thus,
effects of growth temperature tend to plateau between 37oC and 60oC. |
|
Table 1. Parameters of linear regression plots for
relationship of (G+C)% and PLI to optimum growth temperature |
|
.
|
|
.
|
|
(G+C)% versus temperature
|
|
PLI versus temperature
|
|
|
Temperature range |
|
<60o |
60o + |
17o-105o |
|
<60o |
60o + |
17o-105o |
|
Codon Positions
|
.
|
|
Alla |
Y0 |
|
82.5 |
38.6 |
58.5 |
|
-96.1 |
75 |
-15.4 |
|
|
|
|
|
|
|
|
|
|
|
|
Slope |
|
-0.95 |
0.10 |
-0.19 |
|
4.37 |
0.54 |
1.83 |
|
|
Pb |
|
<0.0001 |
0.26 |
<0.0001 |
|
<0.0001 |
0.23 |
<0.0001 |
|
|
|
|
|
|
|
|
|
|
|
|
|
r2 |
|
0.152 |
0.006 |
0.052 |
|
0.160 |
0.009 |
0.211 |
|
|
|
First |
Y0 |
|
78.4 |
51.3 |
62.3 |
|
67.9 |
242.8 |
183.7 |
|
|
|
|
|
|
|
|
|
|
|
|
Slope |
|
-0.65 |
0.02 |
-0.15 |
|
5.86 |
1.23 |
2.20 |
|
P |
|
<0.0001 |
0.731 |
<0.0001 |
|
<0.0001 |
0.088 |
<0.0001 |
|
|
|
|
|
|
|
|
|
|
|
|
|
r2 |
|
0.149 |
<0.001 |
0.063 |
|
0.162 |
0.035 |
0.174 |
|
|
|
Second |
Y0 |
|
55.0 |
39.5 |
45.6 |
|
-154.2 |
22.4 |
-66.3 |
|
|
|
|
|
|
|
|
|
|
|
|
Slope |
|
-0.42 |
-0.03 |
-0.12 |
|
3.61 |
-0.44 |
0.84 |
|
P |
|
<0.0001 |
0.437 |
<0.0001 |
|
<0.0001 |
0.400 |
<0.0001 |
|
|
|
|
|
|
|
|
|
|
|
|
|
r2 |
|
0.131 |
<0.001 |
0.095 |
|
0.086 |
<0.0001 |
0.039 |
|
|
|
Third |
Y0 |
|
114.2 |
25.2 |
67.7 |
|
-202.0 |
-40.2 |
-163.7 |
|
|
|
|
|
|
|
|
|
|
|
|
Slope |
|
-1.77 |
0.32 |
-0.31 |
|
3.64 |
0.84 |
2.45 |
|
P |
|
<0.0001 |
0.087 |
<0.0001 |
|
<0.0001 |
0.246 |
<0.0001 |
|
|
|
|
|
|
|
|
|
|
|
|
|
r2 |
|
0.144 |
0.036 |
0.036 |
|
0.048 |
0.007 |
0.166 |
|
a Data correspond to Figure 1.
b Probability that the slope is not significantly from
zero. For probabilities that slopes are significantly different from each other, please see text.
|
|
The data of Figure 1 are for bases in all codon positions. Table 1 shows that
for some codon positions there is a tendency for progressive effects of growth
temperature at temperatures above 60oC. The third codon position is most
responsive to temperature in the case of (G+C)% (i.e. for the entire temperature
range the slope value of -0.31 for third codon positions is significantly
different from the values of -0.15 and -0.12 for first and second codon
positions; P = 0.024 and 0.007, respectively). In contrast to the trend
at lower temperatures, in the temperature range above 60oC the third codon
position (G+C)% shows a slight increase (a positive slope value of 0.32), which
is of marginal significance (P = 0.087).
On the other hand, the first and third codon positions are most responsive in
the case of the PLIs. For the entire temperature range, the slope values of 2.20
and 2.45, respectively, while not significantly different from each other (P
= 0.43), are both significantly different from the value for second codon
positions (0.84; P<0.0001). For the temperature range above 60oC the
first position PLI tends to continue the positive relationship with growth
temperature observed at lower temperatures (slope 1.23; P = 0.088).
The PLI has contributions from the W bases (ΔW
= A - T) and from the S bases (ΔS = G
- C). These contribute approximately equally to the increase of PLI with optimum
growth temperature. Thus, over the entire range of optimum growth temperatures ΔW
and ΔS values increase with temperature (slopes of 0.88 and 0.95,
respectively, which are not significantly different from each other; P =
0.59; data not shown).
|
|
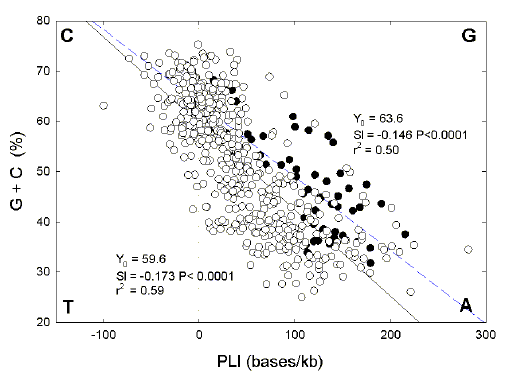
|
|
Fig. 2. Relationship between
(G+C)% and PLI for ORF DNA for 494
prokaryotic species with temperature optima <60oC (open circles; regression
line continuous), and for the 56 species with temperature optima of 60oC and
greater (closed circles; regression line dashed). Single base letters in bold at
the corners indicate quadrants enriched for particular bases [e.g. at high
values of (G+C)% and low values of PLI, there is enrichment for C]. The slopes
of the regression lines are not significantly different (P=0.30).
|
|
As expected from the above, Figure 2 shows that (G + C)% and PLI are
inversely related. Adjusted coefficients of determination indicates that 59% of
the variation in (G+C)% relates to the increase in the PLI in the case of
species with temperature optima below 60oC (open circles), and 50% of the
variation in (G+C)% relates to the increase in the PLI in the case of species
with temperature optima of 60oC and higher (filled circles).
|
|

|
|
Fig. 3. Variation of base density in ORF DNA with the temperature require
for optimal growth of 550 prokaryotic species. (a) The W bases: A, filled
circle; T, open circle. (b) The S bases: G, filled square; C, open square. Data
in the figure show that all slope (Sl) values are significantly different from
zero (P = 0.0009 and less). In addition, slope values for members of each
pair are significantly different from each other in (a) P = 0.0005, and
in (b) P = 0.0001.
|
|
Contributions of individual bases to
ΔW
and ΔS
Individual bases contributed in different ways to the observed changes in
ΔW
and ΔS in response to increased optimum growth temperature. Fig. 3 shows
that the increase in ΔW is derived mainly by an increase in ORF content of
A, while T remains relatively constant. On the other hand, the increase in
ΔS is derived mainly from a decrease in C, while G remains relatively
constant. In essence, as temperature increases, A is traded for C.
In each of Figures 3a and 3b, the regression lines begin to diverge at low
growth temperatures. Thus, at the first codon position, purines are in excess
even at low temperatures, and this difference further increases as optimum
temperature increases. Accordingly, a distinction can be made between a
base-line contribution
to purine-loading, and a
response to a
further pressure to purine-load, in the present case arising from an increase in
growth temperature.
|
|

|
|
Fig. 4. Variation of base density in different codon positions with the
temperature required for the optimal growth of 550 prokaryotic species. Linear
regression parameters shown outside the figures are as in Figure 1. The
probabilities that the slopes of the two lines in each figure are not
significantly from each other are shown within the figure. For other details see
Figure 3.
|
|
Fig. 4 shows that, while A-for-C trading is true for all
codon positions, the major contribution to
ΔW
and ΔS is in the first codon position. However, the major response
to increasing optimum growth temperature is in the third codon position
(greatest positive slope for A, and greatest negative slope for C).
The difference between the slopes for each pair of Chargaff bases is highly
significant in the case of first codon positions (Figs. 4a, d; P<0.0001), and
second codon position S bases (Fig. 4e; P <0.0001). The difference is
significant in the case of third codon positions, but here purines do not exceed
pyrimidines until 63oC (W bases; Fig. 4c) and 71oC (S bases; Fig. 4f). There
is no significant increase in second codon position ΔW
with growth temperature (Fig. 4b), although purines are generally in
excess of pyrimidines. There is symmetry in positive and negative slope values
for the non-Watson-Crick base pairs (A and C; T and G), which is particularly
apparent in third codon positions (e.g. A3, 2.16; C3, -2.14; T3, 0.91; G3,
-0.93).
|
|
Table 2. Average ORF Base Densitiesa at Low or High Growth
Temperatures
|
|
|
|
Base and
Codon
Position
|
|
Optimum Growth Temperature
(degrees Celcius)
|
|
|
|
|
|
|
|
<60b |
|
60+c |
|
Difference |
|
Pd |
|
|
|
A1 |
|
265.7 |
+ or -2.6 |
|
304.3 |
+ or -6.0 |
|
38.6 |
|
<0.00001 |
|
C1 |
|
210.8 |
+ or -2.5 |
|
166.6 |
+ or -5.8 |
|
-44.2 |
|
<0.00001 |
|
G1 |
|
362.5 |
+ or -1.9 |
|
363.9 |
+ or -4.8 |
|
1.4 |
|
0.802 |
|
T1 |
|
161.1 |
+ or -1.5 |
|
165.1 |
+ or -4.5 |
|
4.0 |
|
0.400 |
|
|
|
A2 |
|
304.4 |
+ or -2.3 |
|
325.1 |
+ or -4.5 |
|
20.7 |
|
0.004 |
|
C2 |
|
237.8 |
+ or -1.6 |
|
203.7 |
+ or -3.2 |
|
-34.1 |
|
<0.00001 |
|
G2 |
|
176.6 |
+ or -1.3 |
|
169.3 |
+ or -2.5 |
|
-7.3 |
|
0.073 |
|
T2 |
|
281.2 |
+ or -1.2 |
|
301.9 |
+ or -2.8 |
|
20.7 |
|
<0.00001 |
|
|
|
A3 |
|
194.7 |
+ or -5.5 |
|
260.9 |
+ or -14.0 |
|
66.2 |
|
0.00013 |
|
C3 |
|
309.5 |
+ or -6.6 |
|
244.0 |
+ or -14.8 |
|
-65.5 |
|
0.00127 |
|
G3 |
|
262.8 |
+ or -4.5 |
|
250.9 |
+ or -10.6 |
|
-11.9 |
|
0.390 |
|
T3 |
|
233.0 |
+ or -5.5 |
|
244.1 |
+ or -11.7 |
|
11.1 |
|
0.510 |
|
a Base densities in bases/kb.
b 494 prokaryotic species
c 56 prokaryotic species
d Unpaired t-test.
|
In Table 2 average values for base densities for the 494 species with growth
optima less than 60oC are compared with the corresponding average values for
the 56 species with growth optima of 60oC and higher. First and third codon
positions show major gains in A and declines in C, but small changes in G and T,
which are not statistically significant. For second codon positions the most
significant changes are in the pyrimidines. A significant increase in the purine
A is countermanded by a decrease in the purine G. |
Discussion
|
PLI is positively related to growth temperature
The previously reported positive correlation of
purine-loading with optimum
growth temperature (Lao and Forsdyke 2000) is here confirmed for a much larger
sample of prokaryotes (Fig. 1b). Data from eukaryotes are limited. Our
preliminary studies show that the heat-tolerant marine worm Alvinella
pompejana (Sicot et al. 2000) has much more purine-loading of its mRNA than
comparable mesophilic worms. Among 51 chloroplast genomes, species at the upper
extreme of the distribution of PLI values include the thermophiles Astasia
longa (PLI = 126 bases/kb for 37 sequenced ORFs) and Galeria sulphuria
(PLI 124 bases/kb for 9 sequenced ORFs). However, while the generalization that
thermophiles usually have purine-loaded mRNAs is supported, the converse does
not apply. High purine-loading is found in some mesophilic species (Fig. 1b).
While not excluding other causes of high
purine-loading, such as low
GC-pressure (see below) or an inability to escape usage of purine-rich codons
("protein pressure"), it is possible that purine-loading in non-homeothermic
organisms reflects intermittent exposure to high temperatures. This might apply
to organisms living in the surface layers of tropical soil (e.g. the 24
Clostridial species in GenBank represented by at least 4 ORFs and 2500 bases
have an average PLI of 144 bases/kb).
It should also be noted that pyrexia, an elevation in temperature that
constitutes part of the adaptive response to pathogens (Forsdyke
1995), is
achieved by behavioural adaptation in organisms not able to regulate body
temperature physiologically (e.g. they migrate to full sunlight). If one of the
hosts of a pathogen were non-homeothermic and could elevated its temperature to
high levels behaviourally, then that pathogen might have purine-loaded mRNAs,
many of which might also be expressed in an alternative homeothermic host. In
this light we can interpret high purine-loading in genera such as Borrelia, the
agent of tick-borne infections (8 species in GenBank have an average PLI of 140
bases/kb), and in Plasmodium falciparum (Pizzi and Frontali 2001;
Forsdyke, 2002b; Xue and Forsdyke 2003). In a preliminary study of viruses of
the genus Flavivirus we find non-vector-borne viruses to have less purine-loading than vector-borne viruses, but to show little difference in
(G+C)%
(c.f. Jenkins et al. 2001).
(G+C)% is negatively related to growth temperature
Since rRNAs and tRNAs function by virtue of their structures rather than by
virtue of encoding information for a protein, and since the maintenance of those
structures should be required for function at high temperatures, it is not
surprising that the (G+C)% contents of these RNA species increase with optimum
growth temperature (Forterre and Elie 1993; Dalgaard and Garrett 1993; Galtier
and Lobry 1997). However, mRNAs might not be so constrained. Their structures,
which can appear just as elaborate as those of rRNAs, probably reflect pressures
that operate at the genomic level on both genic and non-genic DNA (e.g. to
assist recombination; Forsdyke and Mortimer 2000; Forsdyke 2001a; Barrette et
al. 2001).
Our observation that ORF
(G+C)% is decreased
in prokaryotes with high
optimum growth temperatures suggests that such genomic operations, even while
requiring secondary structure, can occur at high temperatures without the need
for a (G+C)% increase. This is in agreement with Hurst and Merchant (2001), who
conclude that "within prokaryotes GC content in protein-coding genes, even
at relatively freely evolving sites, cannot be considered an adaptation to the
thermal environment." Similarly, Ream et al.
(2003) note for a variety of
vertebrate species no change in the (G+C)% of specific genes as a function of
average species temperature over the range -1.86oC (fish) to 45oC (desert
reptile). Adaptations to facilitate genomic operations in thermophiles would
include high intracellular concentrations of K+ and other cations,
relaxation of supercoiling, and association with polyamines and specialized
DNA-binding proteins (Forterre and Elie 1993; Stetter 1999; Bernardi 2000;
Forsdyke and Mortimer 2000; Grove and Lim 2001).
Although we assessed the
(G+C)% content only of ORFs, these comprise such a
large part of prokaryotic genomes that our data are likely to reflect total
genomic (G+C)%. However, in contrast to our results (Fig. 1a), values for (G+C)%
derived from buoyant density centifugation and thermal denaturation profiles
demonstrate no correlation between the genomic G+C content and optimal growth
temperature in prokaryotes (Forterre and Elie 1993; Galtier and Lobry
1997). The
latter authors have interpreted their data as supporting a neutralist (non-selectionist) view of the cause of variations in genome base composition at
"freely evolving sites"
(Hurst and Merchant 2001), and as opposing our
proposal that the ubiquitous occurrence of stem-loop potential in genomes is of
adaptive value. Both these interpretations are disputed (Forsdyke and Mortimer
2000; Forsdyke 2002b).
(G+C)% is negatively related to PLI
The general nature of the inverse relationship between PLI and
(G+C)% noted
previously in prokaryotes (Lao and Forsdyke
2000) and eukaryotes (Saccone et al.
2000; Cristillo et al. 2001) is confirmed in the present work (Fig. 2).
Individual codons of a particular amino acid can be differentially affected by
the two pressures. Thus, the frequency of arginine codon CGC increases
progressively with increasing (G+C)%, but the frequency of arginine codon GCA
remains constant, indicating that here the pressure for A to decrease balances
the pressure for G and C to increase (Knight et al.
2001). The latter authors
conclude that "codon and amino-acid usage is consistent with forces acting
on nucleotides
rather than on codons or amino acids"
(our italics;
see Grantham 1980).
Under what circumstances AG-pressure would respond to GC-pressure, or vice
versa, remains to be determined. By the mechanism we have proposed, an
evolutionary chain of causation would be: (i) elevation of temperature; (ii)
increased purine-loading; (iii) decrease of (G+C)%. The latter decrease would be
secondary to the primary adaptation of purine-loading, and hence would be an
indirect adaptation to a thermal environment.
It should be noted that increased AG-pressure per se does not imply a
decrease in (G+C)%. Trading As for Ts would purine-load without affecting (G+C)%.
Trading Gs for Ts would increase the (G+C)%. The decrease implies a preference
for trading As for Cs, perhaps because this would minimize the amino acid
miscoding penalty. Codon flexibility in this respect is most in third codon
positions and least in second codon positions. According to the RNY rule for
average codon composition (Eigen and Schuster 1978; Shepherd
1981), the first
codon position is already R-rich in species with low optimum growth
temperatures, so that, while adept at receiving more A (slope value of 1.26;
Fig. 4a), it is not as adept at receiving more A as is the third codon position,
which is Y-rich in species with low optimum growth temperatures. Thus, A3
responds dramatically to increased optimum growth temperature and has a slope
value of 2.16 (Fig. 4c). Being Y-rich, the third codon position is also
particularly adept at donating C. Thus, C3 has a slope value of -2.14 (Fig.
4f).
The tendency to decrease C in thermophiles would mean there would be less C
at risk of deamination at high temperatures. However, A-for-C trading is also
observed in plots of base composition against (G+C)% for the 1046 prokaryotic
species and 161 bacteriophage species that fulfill our selection criteria (at
least 4 ORFs and 2500 bases in GenBank). For many of these, optimum growth
temperatures are not available; however, as (G+C)% increases, A decreases and C
increases, apparently irrespective of the optimum growth temperature (Mortimer
and Forsdyke 2003).
Nucleic acid constraints on phenotype
Since second codon positions are of major importance in determining the encoded
amino acid, the trading of C2 predicts, for example, that serines (with codons
UCN ) will be replaced in thermophiles by other amino acids, or will be encoded
by purine-loaded codons for serine (AGY). Thus, because the amino acid
composition of thermophile proteins is different from that of mesophile
proteins, it should not be concluded that such changes are necessarily adaptive
with respect to protein function at high temperatures (Lao and Forsdyke 2000;
Lobry and Chessel 2003). Since decreased (G+C)% correlates with an increase in
hydrophilic, charged, amino acid residues (D'Onofrio et al.
1999), and since (G+C)%
and PLI are reciprocally related (Fig. 2), then thermophiles, by virtue of their
high purine-loading, should have proteins rich in charged residues (Lys, Arg,
Glu). These might indeed aid protein stability and militate against protein
aggregation (Jaenicke and Bohm 1998; Cambillau and Claverie
2000), but it should
be noted that the corresponding codons are often purine-rich.
Like GC-pressure (Knight et al.
2001), AG-pressure has the potential to
influence the amino acid content of proteins, and hence protein-dependent
aspects of the phenotype. AG-pressure appears to affect preferentially nucleic
acid segments encoding low-complexity regions of proteins, possibly
surface-located (Fukuchi and Nishikawa
2001). It is proposed that sometimes such
regions exist, not because of a function required at the protein level, but to
permit purine-loading of the corresponding nucleic acid without compromising
protein functional domains. Thus, proteins may be larger than needed for their
function (Pizzi and Frontali, 2001; Cristillo et al. 2001; Forsdyke, 2002b; Xue
and Forsdyke 2003).
Acknowledgements
We thank James Gerlach, Christopher Madill, Andrew Schramm and Scott Smith for
advice and assistance. Jean Lobry and Daniel Chessel kindly made their paper
available prior to publication. Access to the GCG suite of programs was provided
by the Canadian Bioinformatics Resource (Halifax). Academic Press, Cold Spring
Harbor Laboratory Press and Elsevier Science gave permissions for the inclusion
of full-text versions of some of the cited papers in Forsdyke's web pages.
|
|
References
Barrette IH, McKenna S, Taylor DR, Forsdyke DR (2001) Introns resolve the
conflict between base order-dependent stem-loop potential and the encoding of
RNA or protein: further evidence from overlapping genes. Gene
270:181-189
Bell SJ, Forsdyke DR (1999a) Accounting units in DNA. J Theor Biol
197:51-61
Bell SJ, Forsdyke DR (1999b) Deviations from Chargaff's second parity rule
correlate with direction of transcription. J Theor Biol
197:63-76
Bernardi G, Bernardi G (1986) Compositional constraints and genome evolution.
J Mol Evol 24:1-11
Bernardi G (2000) Isochores and the evolutionary genomics of vertebrates.
Gene 241:3- 17
Cambillau C, Claverie J-M (2000) Structural and genomic correlates of
hyperthermostability. J Biol Chem 275:32383-32386
Cantor CR, Schimmel PR (1980) Statistical mechanics and kinetics of nucleic
acid interactions. In: Biophysical Chemistry. WH Freeman, San Francisco, pp. 1183-
1264
Cristillo AD, Mortimer JR, Barrette IH, Lillicrap TP, Forsdyke DR (2001)
Double-stranded RNA as a not-self alarm signal: to evade, most viruses purine-load
their RNAs, but some (HTLV-1, EBV) pyrimidine-load. J Theor Biol
208:475-491
Dalgaard JZ, Garrett A (1993) Archaeal hyperthermophile genes. In: Kates M,
Kushner DJ, Matheson AT (eds) The Biochemistry of Archaea
(Archaebacteria), Elsevier, Amsterdam, pp. 535-562
D'Onofrio G, Jabbari K, Musto H, Bernardi G (1999) The correlation of
protein hydropathy with the base composition of coding sequences. Gene
238:3-14
Eigen M, Schuster P (1978) The hypercycle. A principle of natural
self-organization. Part C. The realistic hypercycle. Naturwissenschaften
65:341-369
Forsdyke DR (1995) Entropy-driven protein self-aggregation as the basis for
self/not- self discrimination in the crowded cytosol. J Biol Sys
3:273-287
Forsdyke DR (1996) Different biological species "broadcast" their
DNAs at different (C+G)% "wavelengths". J Theor Biol
178:405-417
Forsdyke DR (1998) An alternative way of thinking about stem-loops in DNA. A
case study of the G0S2 gene. J Theor Biol
192:489-504
Forsdyke DR (1999) Two levels of information in DNA. Relationship of Romanes'
"intrinsic" variability of the reproductive system, and Bateson's
"residue", to the species-dependent component of the base composition,
(C+G)%. J Theor Biol 201: 47-61
Forsdyke DR (2001a) The Origin of Species,
Revisited. McGill-Queen's
University Press, Montreal
Forsdyke DR (2001b) Functional constraint and molecular evolution. In: Nature
Encyclopedia
of Life Sciences, Vol. 7. Nature Publishing Group, London, pp. 396-403
Forsdyke DR (2002a) Symmetry observations in long nucleotide sequences. Bioinformatics
18:215-217
Forsdyke DR (2002b) Selective pressures that decrease synonymous mutations in
Plasmodium falciparum. Trends Parasitol 18:411-418
Forsdyke DR, Mortimer JR (2000) Chargaff's Legacy. Gene
261:127-137
Forterre P, Elie C (1993) Chromosome structure, DNA topoisomerases, and DNA polymerases in archaebacteria (archaea). In: Kates M, Kushner DJ, Matheson AT
(eds) The Biochemistry of Archaea (Archaebacteria). Elsevier, Amsterdam, pp.
325-345
Fukuchi S, Nishikawa K (2001) Protein surface amino acid compositions
distinctively differ between thermophilic and mesophilic bacteria. J Mol Biol
309:835-843
Galtier N, Lobry JR (1997) Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes.
J Mol Evol 44:632-636
Grantham R (1980) Workings of the genetic code. Trends Biochem Sci
5:327-331
Grove A, Lim L (2001) High affinity DNA binding of HU protein from the
hyperthermophile Thermotoga maritime. J Mol Biol
311: 491-502
Hurst LD, Merchant AR (2001) High guanine-cytosine content is not an
adaptation to high temperature: a comparative analysis among prokaryotes. Proc R Soc Lond B
268:493-497
Jaenicke R, Bohm G (1998) The stability of proteins in extreme environments.
Curr Opin Struct Biol 8:738-748
Jenkins JM, Pagel M, Gould EA, Zanotto PM de A, Holmes EC (2001) Evolution of
base composition and codon usage bias in the genus Flavivirus. J Mol Evol
52:383-390
Knight RD, Freeland SJ, Landweber LF (2001) A simple model based on mutation
and selection explains trends in codon and amino-acid usage and GC composition
within and across genomes. Genome Biol. 2:0010.1-0010.13.
Lao PJ, Forsdyke DR (2000) Thermophilic bacteria strictly obey Szybalski's
transcription direction rule and politely purine-load RNAs with both adenine and
guanine. Genome Res 10:228-236
Lauffer MA (1975) Entropy-Driven Processes in
Biology. Springer-Verlag, New
York
Lobry JR, Chessel D (2003) Internal correspondence analysis of codon and
amino-acid usage in thermophilic bacteria. J Appl Genet
44:235-261.
Mortimer JR, Forsdyke DR (2003) Comparison of responses by bacteriophage and bacteria to pressures on the base composition of open reading frames.
Appl. Bioinformatics 2:47-62.
Nakamura Y, Gojobori T, Ikemura T (2000) Codon usage tabulated from the
international DNA sequence databases: status for the year 2000. Nucleic Acids
Res 28:292
Pizzi E, Frontali C (2001) Low-complexity regions in Plasmodium falciparum
proteins. Genome Res. 11:218-229
Ream RA, Johns GC, Somero GN (2003) Base compositions of genes encoding
α-actin and lactate dehydrogenase-A from differently adapted vertebrates show no
temperature-adaptive variation in G + C content. Mol Biol Evol
20:105-110
Saccone C, Gissi C, Lanave C, Larizza A, Pesole G, Reyes A (2000) Evolution
of the mitochondrial genetic system: an overview. Gene
261:153-159
Shepherd JCW (1981) Method to determine the reading frame of a protein from
the purine/pyrimidine genome sequence and its possible evolutionary
justification. Proc Natl Acad Sci USA
78:1596-1600
Sicot F-X, Mesnage M, Masselot M, Exposito J-Y, Garrone R, Deutsch J, Gaill F
(2000) Molecular adaptation to an extreme environment: origin of the thermal stability
of the Pompeii worm collagen. J Mol Biol
302:811-820
Smithies O, Engels WR, Devereux JR, Slightom JL, Chen S-h (1981) Base
substitutions, length differences and DNA strand asymmetries in the human Gg
and Ag fetal
globin gene region. Cell 26:345-353
Stetter KO (1999) Extremophiles and their adaptation to hot environments. F E
B S Lett 452:22-25
Szybalski W, Kubinski H, Sheldrick O (1966) Pyrimidine clusters on the
transcribing strand of DNA and their possible role in the initiation of RNA synthesis.
Cold
Spring Harb Symp Quant Biol 31:123-127
Xue HY, Forsdyke, DR (2003) Low complexity segments in Plasmodium
falciparum proteins are primarily nucleic acid level adaptations. Mol.
Biochem. Parasitol. 128:21-32
|

End-Note 2004
Some of the main findings in
this paper were confirmed by Paz et al. (2004;
Adaptive role of increased
frequency of polypurine tracts in mRNA sequences of thermophilic prokaryotes.
Proc.
Natl. Acad. Sci. USA 101, 2951-2956). They also found that heat-shock protein mRNAs had a much higher R/Y
ratio than most other mRNAs, both in thermophiles and mesophiles, and in five
eukaryotic species.

End-Note 2005
In contrast to the results of ourselves and others, Musto
and coworkers [FEBS
Lett. 573
(2004) 73]
reported that positive correlations are generally
observed between GC levels and optimal growth temperatures in 20 prokaryotic
families, and many of these are significant. However, Sayed-Amir Marashi
and Zahra Ghalanbor quickly showed (Biochem.
Biophys. Res. Comm. 325, 381-3)
that these correlations are not
robust, i.e., "correlation coefficients and/or significance of correlations
could be considerably influenced by exclusion of very few (even as small as one) species from each dataset."
Apparently unaware of the work described here, they cautiously concluded that solely based on
Musto's data, "one cannot establish that GC contents of prokaryotic genomes increase as a result of growth temperature increments."
The issue was further considered in 2005 in subsequent issues of
Biochem.
Biophys. Res. Comm.(e.g. Musto et
al. 330, 357-360 and Basak & Ghosh, 330, 629-632).

End-Note
January
2007
In
a paper entitled "Protein and DNA Sequence Determinants of Thermophilic
Adaptation" Zelfovich, Berezofsky and Shakhnovich (January 2007.
PLOS
Computational Biology 3, issue 1,
62-71) declare as the "key question": "Which factor - amino acid
or nucleotide composition - is primary in thermal adaptation and which is
derivative?" They consider previous evidence on this as
"anecdotal" and claim to present evidence for "an exact and
conclusive" relationship, based on an "exhaustive study" that
provides a "complete picture." A set of amino acids - IVYWREL - gives
the best correlation with optimum growth temperature. They note:
"Signatures
of thermal adaptation in protein sequences can be due to the specific
biases in nucleotide sequences and vice versa. - - One has to explore
whether a specific composition of nucleotide (amino acid) sequences
shapes the content of amino acid (nucleotide) ones, or thermal
adaptation of proteins and DNA (at the level of sequence compositions)
are independent processes." |
Conclusion:
|
"Resolving
the old-standing controversy, we determined that the variation in
nucleotide composition (increase of purine-load, or A + G content with
temperature) is largely a consequence of thermal adaptation of
proteins." |
The
superficial reader of the paper, while noting the purine-richness of some of the
codons corresponding to the IVYWREL amino acids, will perhaps conclude that the
"independent processes" alternative has been excluded. Reading the
paper (e.g. their Figure 7) one can question the validity of this conclusion.
Many of the IVYWREL amino acids have purine-poor alternative codons (especially
IYLV, which at best can only change one purine unit in their codons). One of the
IVYWREL amino acids have relatively purine-rich alternative codons (R, which at
best can change two purine units). Two (EW) are always purine-rich, and there
are no alternatives.
Thus, displaying more EW's as the
temperature got hotter would satisfy a need both for more purines and for more
tryptophan and glutamate, so here there is no discrimination as to whether one
"shapes" the organisms's content of the other. Displaying more IYLVs
gives only minimal flexibility in accommodating a purine-need. Most flexibility
is provided by R (arginine) codons. The authors do not give statistics for the
differences between the slopes of Figs. 7a (unshuffled codons) and 7b (shuffled
codons), but they appear real, presumably reflecting the choice biologically of
purine-rich codons, a choice the organisms might not have to make if there were
no independent purine-loading pressure. Thus, the authors note, but only in
parenthesis, that the slopes "are somewhat different suggesting that
codon bias may be partly responsible for the overall purine composition of
DNA." Given the boldness of their conclusions, this is somewhat
startling.
An analogy may help. Passing from winter
to summer, you change your outdoor attire. Many people (Ismelda Marcos excepted)
have a wider range of shirts and sweaters than of footwear. There may be items
which provide more reliable indices of outside temperature than others. For
example, the weight of your shirt + sweater might more finely correlate with
temperature than the weight of your shoes. If you wanted to predict outside
temperature from attire, you would choose shirt-sweater weights, rather than
shoe weights. But if your clothes (shirt + sweater) weigh more, you might need
sturdier shoes to cope with the extra weight. Thus, shoe-weight would depend on
clothes-weight. The latter would be primary and the former would be secondary
("derivative"). But showing a better correlation with temperature in
the case of shirt + sweater, does not establish a dependence of shoe-weight on
shirt-sweater weight. More likely, you wear lighter shoes in warm weather
because they keep your feet cooler! Both depend on temperature. It just so
happens that the correlation with temperature is coarser and not so finely tuned
for shoe weight, as for shirt-sweater weight.

End-Note August 2012
Dehouck et al. (2008) report that relationship
between a protein's in vitro thermostability and the optimum growth
temperature of the organism containing it, is not so close as previously
thought. Furthermore, Liu et al. (2012) now conclude from a
study of xylanase purine-rich coding sequences that "The codons relating
to enzyme thermal property are selected by thermophilic force at [the]
nucleotide level," not at the protein level.
Dehouck Y, Folch B, Rooman M (2008) Revisiting the
correlation between proteins' thermoresistance and organisms'
thermophilicity. Protein
Engineering, Design and Selection 21,
275-278.
Liu L, Wang L, Zhang Z, Wang S, Chen H (2012)
Effect of codon message on xylanase thermal activity.
Journal of Biological Chemistry
287, 27183-27188.
End-Note August 2015
Maria
Brbic et al. (2015) report that oligonucleotide frequency is a major force
to which protein pressure is often subservient. The emergence of the
PubMed comment facility allows commentary on papers in journals that
normally have not allowed this. Thus, I have commented positively there.
Brbic M, Warnecke T, Krisko A, Supek F (2015)
Global shifts in genome and proteome composition are very tightly coupled.
Genome Biology and Evolution
7, 1519-1532.
|



