The selfish gene revisited:
reconciliation of Williams-Dawkins and
conventional definitions
Donald R. Forsdyke
![]()
Darwin_missed_Mendel,_not_Sanderson_and_Beale
Gene,_not_individual,_as_unit_of_heredity
Agendas,_trade-offs,_and_kin-selection
Structure-mediated_homology_recognition
Strand_migration_to_the_genic_boundary
End_Note_on_Hotspots_(November_2011)
End_Note_on_Akiyoshi_Wada_(October_2014)
![]()
ABSTRACT Sightings of the revolutionary comet that appeared in the skies of evolutionary biology in the 1976 - the selfish gene - date back to the nineteenth and early twentieth century. It became generally recognized that genes were located on chromosomes and compete with each other in a manner consistent with the later appellation "selfish." Chromosomes were seen as disruptable by the apparently random "cut and paste" process known as recombination. But each gene was only a small part of its chromosome. On a statistical basis a gene should escape disruption for many generations. This led George Williams and Richard Dawkins to a new definition of the gene differing from conventional biochemical definitions in that there were no consistent genic boundaries. There had been no previous sightings of another revolutionary, albeit less verbally spectacular, comet that appeared in 1975 - the homostability principle of Akiyoshi Wada. Each gene has a base composition "accent" that distinguishes it from its neighbours. We now see that recombination can be triggered by the shift in base composition at genic boundaries. Hence, the Williams-Dawkins definition approaches the conventional. |
Keywords:
Base composition, Chromosome, Crossing-over, Gene definition,
Homostability, Meiosis, Recombination
![]()
Darwin missed Mendel, not Sanderson and Beale
Although Mendel's famous paper was published in
1866, its importance was not recognized until 1900 when the Mendelian
revolution began (Cock and Forsdyke 2008). But another 1866 publication
was brought to Charles Darwin's attention by an article in
Gardeners' Weekly (Beale
1866; Darwin 1868: 378). This was a Royal Commission Report on the
cattle plague - now known to be caused by a virus - that was then
ravaging
Another contributor, the pathologist Lionel
Beale, concluded: "With regard to the nature of the contagium itself,
evidence has been adduced to show that it consists of very minute
particles of matter in a living state, each capable of growing and
multiplying rapidly when placed under favourable conditions." In earlier
testimony to the Commission, the Medical Officer to the Privy Council
had declared (Simon 1865):
|
Hourly observation tells us that the contagium
of smallpox will breed smallpox, that the contagium of typhus will breed
typhus, that the contagium of syphilis will breed syphilis, and so
forth, - that the process is as regular as that by which dog breeds dog,
and cat cat, as exclusive as that by which dog never breeds cat, nor cat
dog. |
These observations, which extended Louis
Pasteur's work on the, then highly controversial, germ theory of disease
(O'Malley 2009), led to Darwin's
"provisional hypothesis of pangenesis" (Darwin 1868: 357-404; Forsdyke
2001). He proposed that the individual characters of an organism could
be transmitted through the generations in the form of minute "gemmules"
- now best equated with genes. These were "capable of largely
multiplying themselves by self-division, like independent organisms" and
were part of the "formative matter which is contained within the
spermatozoa." Thus, "the child, strictly speaking, does not grow into
the man, but includes germs which slowly and successively become
developed and form the man."
Given that a
"contagium" could spread from
animal to animal, it was easy to believe that a gemmule, after
appropriate somatic education, might leave its cell of origin and
transfer to the germ line, so that parental experience could be
transmitted to offspring. While this Lamarckian aspect of pangenesis
gained little support, the idea that the "gemmules," or "pangens," could
correspond to individual characters, was deemed sound by Hugo de Vries
(1889).
Darwin's mathematician son, George, had
calculated the minute size of the inorganic molecules that chemists were
then studying and Darwin noted (Darwin 1875: 373-374): "No doubt the
[organic] molecules of which an organism are formed are larger, from
being more complex, than those of an inorganic substance, and probably
many molecules go to the formation of a gemmule; but -- we can see what a
vast number of gemmules -- [an organism] might contain." He concluded
that: "Each living creature must be looked at as a microcosm - a little
universe, formed of a host of self-propagating organisms, inconceivably
minute and numerous as the stars in heaven" (Darwin 1868: 404).
Although unaware of Mendel, many biologists at
that time knew that in crosses between dissimilar types certain
characters (e.g. tallness in peas) were dominant ("prepotent") over
others (e.g. smallness in peas; Roberts 1929: 170). When considering
prepotency,
|
It is a probable hypothesis, that what the world is to organisms in general, each organism is to the molecules of which it is composed. Multitudes of these, having diverse tendencies, are competing with each other for opportunities to exist and multiply; and the organism, as a whole, is as much a product of the molecules which are victorious as the Fauna or Flora of a country is the product of the victorious beings in it. On this hypothesis, heredity transmission is the result of the victory of particular molecules contained in the impregnated germ. Adaptation to conditions is the result of the favouring of the multiplication of those molecules whose organizing tendencies are most in harmony with such conditions. |
More colourfully,
|
Protoplasm is, for all its structurelessness,
composed of an infinite number of living molecules, each of them with
hopes and fears of its own, and all dwelling together like Tekke
Turcomans, of whom we read that they live for plunder only, and that
each man of them is entirely independent, acknowledging no constituted
authority. |
The case was made more formally by the
embryologist Wilhelm Roux (1881). To the end of his life, Huxley (1893:
vi) persisted in his belief in a "struggle for existence within the
organism."
Although not agreeing with the Lamarckian
aspects of his cousin's pangenesis hypothesis, Francis Galton accepted
that an organism could be viewed as a constellation of characters for
each of which there could be a corresponding gemmule, which he referred
to as an "element." In
|
As we know nothing about the arrangements and
movements of the ultimate living units or germs, we can answer only by
analogies. The exact answer would require a knowledge of the cause of
what, in the nomenclature of Weismann, would be called the architecture
of the id [structure of the
gene], and of which he assumes the existence, but does not attempt to
account for. We know that the germ contains the seeds of a vast number
of ancestral potentialities, only a very few of which can be
simultaneously developed, being to a great extent mutually exclusive. It
may therefore be inferred with confidence, that organisation is reached
through a succession of struggles for place among competing elements,
the successful ones owing their success through position, through
superiority in vigour, and so on (Galton's italics). |
His case was broadly framed in abstract terms:
| However vague such an explanation may be, it is far from
being an inefficient one, for it defines the general character
of a process though avowedly incapable of dealing with the
details. It applies, moreover, to every theory of heredity which
is of a 'particulate'
character; - that is to say, wherever the theory is based on the
supposition of a vast number of partly independent biological particles
-- . Theories that have this general idea for their foundation seem to be
the only ones that are in any way defensible (Galton's italics). |
From this we see that, in apparent ignorance of
Mendel's work, by 1899 the idea of what we now refer to as "genes," with
a degree of competitiveness that we can now refer to as "selfish," was
established in some influential quarters. Although when considering sex
ratios in 1871
Gene, not individual, as unit of heredity
Following the discovery of Mendel's work in
1900, the zoologist William Bateson and the horticulturalist Charles
Hurst became its major advocates in the English-speaking world (Cock and
Forsdyke 2008). Their elliptical writings, especially those of Bateson,
made heavy demands on readers. For example, the word "gene" having not
been yet coined, rather than Galton's "elements" or Weismann's "ids"
they referred to "factors" - a term that had to be carefully interpreted
in context. Parental gametes united to form a zygote that grew into an
adult that then produced fresh gametes. This process was fundamentally
the same in plants and animals. It was now recognized that, during
gametogenesis, each pair of genes ("allelomorphs" corresponding to a
particular character) that had been separately introduced into the
zygote with each parental gamete, were again separated ("segregated").
Having noted Michael Guyer's description of the
cytology of spermatogenesis (Bungener and Buscaglia 2003), on October 1st
1902 Bateson (1904) declared that there was "reason to believe that the
chromosomes of the father plant and mother plant, side by side,
represent blocks of parental characters". But it took the next two
decades to clarify the cytological details, which were elaborated as the
"Sutton-Bovari" hypothesis by Edmund Wilson (1925). During meiotic
division in the gonads, homologous chromosomes were seen to pair and
randomly exchange opposing segments ("recombination"). There was "crossing over"
- a breaking and joining between the chromosomes.
This meant that, while order was usually unchanged, the set of the
segments in each chromosome of an emerging gamete (today referred to as
its "haplotype") was different from that of the corresponding
chromosomes of the parent.
In an address to the Leicester Literary and
Philosophical Society Hurst (1904) referred to germ cells (gametes) as
containing a "factor of" or "factor for" a character,25 and
added:
| Seed shape and seed colour are distinct characters with an independent inheritance. Mendel's discovery of this elementary fact was one of the secrets of his success, and his experimental demonstration of the existence of single hereditable characters proved once and for all that the true unit of heredity is not the individual [organism], but the [factor for a] single character (Hurst's italics). |
Guided by the interpolations within square
brackets, the modern reader can see that, if by "the true unit of
heredity"
Bateson (1919) expanded on this in an address to
the Yorkshire Natural Science Association on the "mongrel" composition
of the populations of modern nation states:
|
The fact however, which I wish more to
emphasize, is that by the workings of the phenomenon of genetic
segregation a man's children may possess few of the transferable
ingredients [genes] which characterized him, his grandchildren may
possess none at all, and of his collaterals it is practically certain
that few will contain so much of his that he need feel any personal
satisfaction or humiliation in their performances. -- Looked at coldly in
the light of physiological knowledge, what is called the tie of blood is
therefore in modern times exceedingly slender, and in all likelihood
many of us contain no more of the elements that went to the making of
Shakespeare and our heroes than the modern Greek contains of Zeus or
Phoebus, despite the frequent alliances which those deities contracted
with the daughters of man. |
That a man might transfer
no genes to his
grandchildren, although consistent with some of the observations of
Guyer, somewhat overstated the case. In "some remarks about units of
heredity" Wilhelm Johannsen (1923), whose first language was not
English, spelled out the biological detail:
|
Gametogenesis with chromosome-reductions,
accompanied by reformations and, as it were, partial rejuvenescence of
cell-structures, must in some way act as if especially organized for
obliterating the individual's
personally 'acquired characters,' which as a rule totally disappear in
sexual reproduction. -- Continuity
in inheritance, the cardinal idea of Aristotle, is - as applied to
Mendelian heredity - represented by the continuity of chromosomes in the
forthcoming generations - but [is] greatly complicated by disjunctions
and recombinations of chromosome pairs. This hereditary continuity is --
dissolved into -- regular periodic discontinuities: Mendelian heredity
always operates with discreet genotypical elements [genes]. Hence
differences are here always discontinuous -- chemical constitutional
differences. Phenotypes however may show discontinuous as well as all
degrees of continuous variation (Johanssen's italics). |
Bateson's death in 1926 marked the end of an
era. By then the word "gene," introduced by Johanssen around 1909, was
well established, as was the notion that genes were distributed along
chromosomes in linear order. Sexual reproduction, through recombination
between homologous chromosomes, was seen as disrupting that order so
that, seeming to be the least disruptable, it was the gene that appeared
to be the most stable unit of heredity (Morgan 1926). Although the idea
of competition was around, the extension of this, to the idea that a
gene might have its own agenda, was for the future. But it was the close
future, not the distant future.
Agendas, trade-offs, and kin-selection
In the 1920s work on pneumococcal transformation initiated studies
leading to the biochemical characterization of genes (Griffith 1928;
Olby 1974). Shortly thereafter, the polymath J. B. S. Haldane (his
initials being derived from his above-mentioned uncle, John Burdon Sanderson) united two apparently disparate concepts. First, since
plant fertilization involved transfer of the male germ by way of the
pollen tube to the ovule, then if multiple pollen grains alighted on the
female stigma, success would go to the grain whose tube grew fastest.
Second, a single gene expressed at two different stages of a life cycle
might produce different, stage-specific, outcomes (i.e. it could be
pleiotrophic); ideally, it would be beneficial at both stages, but a
mutant form of the gene might benefit one stage, not the other
(antagonistic pleiotrophy). Haldane (1932) wrote:
|
A higher plant species is at the mercy of its pollen grains. A [mutant]
gene which greatly accelerates pollen tube growth will spread through a
species [enhance its own survival] even if it causes moderately
disadvantageous changes in the adult plant. [Conversely] a gene
producing changes which would be valuable in the adult would be unable
to spread through a community if it slows down pollen tube growth. |
The ability of a gene to influence fertilization is likely to dominate,
since without fertilization there can be no adult. On the other hand, a
less than perfect adult might still be able to reproduce sufficiently to
ensure the gene's passage to future generations where it would again
promote fertilization at the expense of adult fitness. Thus, individual
plants can be at the mercy of a mutant gene in pollen grains and, in the
absence of any countervailing influence, eventually the gene should
spread to the entire species. In the apparently ascending hierarchy -
gene, organism, species - it would be the "agenda" of the lowest member
that was followed.
Haldane also noted that genes that promoted altruistic conduct would be
included in this category: "For in so far as it makes for survival of
one's descendants and near relations, altruistic behaviour is a kind of
Darwinian fitness, and may be expected to spread as the result of
natural selection." This idea was mathematically elaborated in terms of
trade-offs and preferential kin-selection by William Hamilton (1964). At
that time George C. Williams was composing his seminal text
Adaptation and Natural Selection
(Williams 1966). These two - Hamilton and Williams - were the main
antecedents acknowledged by Dawkins (1976).
It follows from the mechanics of recombination
that genes, rather than the chromosomes that contain them, are most
likely to remain intact through the generations. Thus children differ
from their parents because they have different combinations of genes,
but a given gene in a child is usually identical to that of the parent
it was inherited from. This theme, set out by Bateson and Johannsen
above, was echoed by Williams. However, he went further in proposing
that genes should be defined
by this characteristic (Williams 1966: 22-25):
|
Socrates' genes may be with us yet, but not his
genotype, because meiosis and recombination destroy genotypes as surely
as death. It is only the meiotically dissociated fragments of the
genotype that are transmitted in sexual reproduction, and these
fragments are further fragmented by meiosis in the next generation. If
there is an ultimate indivisible fragment it is, by definition, 'the
gene' that is treated in the abstract discussions of population
genetics. -- I use the term gene to mean 'that which segregates
and recombines with appreciable frequency.' Such genes are potentially
immortal (William's italics). |
Dawkins (1976) agreed:
|
Sexual reproduction has the effect of mixing and
shuffling genes. This means that any one individual body is just a
temporary vehicle for a short-lived combination of genes. The
combination of genes which is
any one individual may be short-lived, but the genes themselves are
potentially very long-lived. Their paths constantly cross and re-cross
down the generations. One gene may be regarded as a unit which survives
through a large number of successive individual bodies (Dawkins'
italics). |
We must here digress to note that for present
purposes we do not follow Williams (1985) in his view that "the gene is
not the [DNA] molecule, but the information coded by the molecule". This
was later elaborated in Natural
Selection: Domains, Levels, and Challenges
where he noted (Williams 1992:
10-13) that: "Information can exist only as a material pattern, but the
same information can be recorded in a variety of patterns in many
different kinds of material. A message is always coded in some medium,
but the medium really is not the message." Here Williams conflates the
words "message" and "information." This is easily done. For example, "Did you get his message?" can enquire either whether you received the
medium (e.g. paper in an envelope), or whether you had understood the
information contained in that medium (i.e. whether information
flowed from the medium to
your head). Here we take the medium to be the message (McLuhan 1964). A
DNA sequence (gene), if subjected to an appropriate reading process, can
be seen to contain
information (e.g. for the amino acid sequence of a protein). But the
information itself is an abstract entity. The medium - a sequence of
bases in DNA - is the
message. This message contains
information, but is not itself
that information. Genes are DNA! Typically, information
flows from DNA (gene), by way
of messenger RNA (not a gene, even though containing genic information),
to protein. This particular form of information - protein-encoding
information - is but one of many
forms of information that can be contained simultaneously in a given DNA
sequence (Forsdyke 2006: 183-224). These forms include "accent" or
"dialect" (see below).
Although certain parts of genomes were known to
be more prone to recombination than others ("hotspots"), it was not
thought that recombination would respect genic boundaries. Recombination
occurred both between and
within genes. While averring
that "a body is the genes' way of preserving the genes unaltered,"
Dawkins (1976) recognized that William's "ultimate indivisible fragment"
might need some qualification:
| Even a cistron [gene] can be cut in two by crossing over. The gene is defined as a piece of a chromosome which is sufficiently short for it to last, potentially, for long enough for it to function as a significant unit of natural selection (Dawkins' italics). |
Thus, to get round the boundary problem, Dawkins
advanced a statistical conception of the gene. Genes were small parts of
genomes, so the probability was high that a small chromosome segment
containing a gene would escape meiotic fragmentation for many
generations. While eventually the gene would succumb to the
recombination mill, he implied that this would be a rare event:
|
Individuals are not stable things, they are
fleeting. Chromosomes too are shuffled to oblivion, like hands of cards
soon after they are dealt. But the cards themselves survive the
shuffling. The cards are the genes. The genes are not destroyed by
crossing-over, they merely change partners and march on. |
The perspective of Williams and Dawkins was very
different from that of many biochemists who had to deal with real DNA
sequences, and had to associate gene names with distinct segments for
which start and stop signals had to be precisely assigned (Forsdyke
2009; Griffiths and Stotz 2006; Stotz 2009). In extreme form, the
Williams-Dawkins definition would include an entire chromosome as a "gene" if that chromosome happened to be excluded from recombination (as
in the case of chromosomes in the male fruit fly germ line). So should
we shrug off the Williams-Dawkins gene definition in terms of
recombination as rhetorical, designed to emphasize their new
perspective, rather than as a serious attempt to delineate the gene? Or
could they, unknowingly, have been pointing to a hidden genic boundary,
somehow related to recombination? Here we turn to the homostability
principle and its role in recombination.
The revolutionary
selfish gene "comet" that
appeared in the skies of evolutionary biology in 1976 was widely
observed. It followed a no less important, but largely unobserved,
homostability principle "comet" that had appeared in 1975. Each gene,
while adhering to the general trend in base composition of the genome
that contains it, has its own distinctive base composition - it has a
distinctive proportion of the four DNA bases that are either G or C
(i.e. a distinctive "GC%"). The biophysicist Akiyoshi Wada, whose first
language was not English, described this as a "homostabilizing
propensity" that might relate to recombination (Wada et al. 1975):
|
Genetic information is
'written' by a variation
in sequence on the one hand, and the physical stability of the
double-stranded structure is determined by the base composition on the
other hand. -- DNA is found to consist of a number of homostability
regions which come from homogenous base sequences consisting of 500 base
pairs or more. -- Biologically, it is hard -- to believe that such
regional homostability originates in a fundamental characteristic of the
genetic code itself. It is quite plausible -- that the homostability
region plays an important part somewhere in the biological process
within which the DNA is closely related. If so, then the evolutionary
selective force can be considered to have fixed such regions of DNA.
From the size of the homostability region, recombination might be one
possible process which is aided by it. In any cases, the wobble bases
[in codons] must give the necessary redundancy to make a homostability
region without spoiling the biological meaning of the genetic code. |
There had already been theoretical
pronouncements by Holliday (1968) and Schaap (1971) that there were at
least two hierarchical levels of information in DNA, one of which might
specially relate to recombination. But, to this theory, Wada and
coworkers added hard data. They distinguished base order-dependent "genetic information" that, in the tenor of their times, largely meant
protein-encoding information, and base composition-dependent "genetic
information." They recognized that "to make a homostability region"
other functions might be threatened, but potential conflicts could be
minimized "without spoiling the biological meaning" through the "necessary redundancy" of the genetic code (e.g. to encode glycine, a
low GC% gene uses GGT or GGA, and a high GC% gene uses GGC or GGG).
The emergence of sequencing technologies in the
1970s permitted the boundaries of the Wada homostability regions to be
approximated to those of genes (Bibb et al. 1984; Wada and Suyama 1986),
and the term "microisochore" was suggested (Forsdyke 2004). This
indicated a region of uniform base composition less extensive than the
large sub-genomic segments of uniform base composition ("isochores")
that Georgio Bernardi and his associates had described (Filipski et al.
1973).
The work of Erwin Chargaff in the 1950s had
shown that biological species tended to differ in their overall genomic
GC% values (Chargaff 1963). Indeed, in 1980 Richard Grantham advanced
his "genome hypothesis," noting that a distinctive base composition
could be viewed as the "dialect" or "accent" of a genome that would be
imposed on its genes (Grantham 1980; Grantham et al. 1986; Paz et al.
2006). Yet, within this genome framework there can be further "accent"
differentiation. Thus, today we can distinguish three levels of
homostability (i.e. GC% uniformity) - microisochores (genes), "macroisochores" (isochores), and whole genomes. We here omit
consideration of macroisochores.
Structure-mediated homology recognition
Central to the issue is the homology search
process by which nucleic acid molecules recognize each other prior to
recombination, a process that can occur both in somatic cells and when
homologous chromosomes pair during meiosis in germ-line cells. Despite
decades of research on recombination and its associated enzymes, it is
lamented that "the mechanism by which homologs uniquely pair with each
other is poorly understood" (Blumenstiel et al. 2008). We still have
little idea how two "homologous needles find each other in the genomic
haystack" (Barzel and Kupiec 2008). We are urged "to 'branch out' in our
thinking about meiotic recombination" (Cromie and Smith 2007).
Homologous recombination occurs between
sequences which are identical, or very closely so. It is easy to imagine
- and many textbook diagrams support the idea - that first one strand of
a DNA duplex is cut and then a free single-strand seeks a complementary
strand in another DNA duplex. However, certainly in fruit fly and
nematode worms, homology recognition does not require initial strand
breakage (Moore and Shaw 2009). Indeed, homology recognition can occur
between intact DNA duplexes in a simple salt solution in the absence of
proteins (Kornyshev and Wynveen 2009; Danilovitz et al. 2009).
As to the mechanism, there is evidence that
"the
DNA duplex is labile on a millisecond time scale, allowing local bubble
formation" (i.e. strand separation without breakage), so that "alignment
of DNA molecules of identical sequence and length could be stabilized by
such perturbations occurring simultaneously (and transiently) at
identical positions in the sequence" (Inoue et al. 2007). This has been
referred to as "structure-mediated homology recognition" (Baldwin et al.
2008; Kornyshev 2010). There is some, albeit indirect, evidence
consistent with this occurring in
vivo without participation of gene products (i.e. RNA and proteins;
Bateman and Wu 2008; Blumenstiel et al. 2008).
But the "bubbles" would be unlikely to remain
single stranded. If not prevented by single-strand binding proteins or
unusual sequence characteristics, the extruded single-strands should
collapse on themselves and adopt folded stem-loop configurations. In
studies of pairing between single-stranded RNA molecules, Tomizawa
showed that complementary molecules must first recognize each other,
prior to hybridization, by
exploratory "kissing" interactions (base pairing) between the loops of
stem-loop structures (Eguchi et al. 1991). By analogy, Kleckner and
Weiner (1993) proposed that this might apply to the pairing of
complementary DNA sequences. Initial loop-loop interactions would
progress to the annealing of complementary strands (Figure 1), thus
setting the stage for strand breakage and recombination.
Recombination being a genome-wide activity, this proposal led to
predictions that could be subjected to bioinformatic analysis:
(i) the
potential to extrude stem-loops would be widely distributed; (ii) a
regularity in DNA base composition, known as Chargaff's second parity
rule ("PR2"), would tend to apply generally to single-strands of DNA.
Both predictions were confirmed (Forsdyke 1996). For example, consider
the following two stranded DNA sequence where a T in the top strand
pairs with an A in the bottom strand (and vice versa), and a C in the
top strand pairs with a G in the bottom strand (and vice versa). Stare
at the sequence as long as you like and nothing remarkable is likely to
emerge:
|
TGCGACGCCGATAGCGTCGCA |
|
Yet, the sequence has a special property found widely distributed in biological DNA sequences. If we peel away the top strand it can then fold into the following form with two major elements, a stem and a loop:
C
TGCGACGC G
ACGCTGCG TA
A
For
this structure to form there have to be appropriately arranged
("palindromic") sets of matching (complementary) bases. The stem
consists of paired bases (T matching A, and C matching G). Only the
bases in the loop (CGATA)
are unpaired. If you count the bases,
A=4,
C=7,
G=7 and
T=3. Numerically T
approximates to A, and C approximates to G, so PR2 applies.
Some thought that PR2 would be explained in
terms of "mutational biases" that were of little biological significance
(Sueoka 1999). However, it is now agreed that genomes contain sequences
that "may be under selective pressure to preserve their palindromic
character and therefore follow PR2 (as pure palindromic sequences are
effectively base paired)." (Lobry and Sueoka 2002). Indeed, Bultrini et
al. (2003), noting a "symmetrical trend" in DNA sequences, invoked
"formation of stem-loop structures."
From this perspective, some long-standing
recombination models, classified as "paranemic" (no initial strand
breakage), can be seen as having much to commend them (Crick 1971;
Sobell 1972; Wagner and Radman 1975; Doyle 1978; Wilson 1979). Evidence
for "paranemic crossover DNA" is mounting (Wang et al. 2010). Its
importance for the present paper is that the rate-limiting step in
recombination is likely to be, not the actual
pairing of homologous DNA
strands, but the extrusion of
appropriate stem-loop structures from duplex DNA (Figure 1). The
extrusion is likely to be symmetrical, affecting both strands of a
duplex equally, and to be critically dependent on base composition
(Forsdyke 2007). When the GC% values of two sequences are close, the
shapes and tempos of stem-loop extrusions can be similar, a condition
propitious for the strand pairing that precedes recombination. When GC%
values differ, then, however similar sequences are in other respects,
the pairing is disfavoured. Asymmetrical extrusion is considered
elsewhere (Lao and Forsdyke 2000; Zhang et al. 2008).
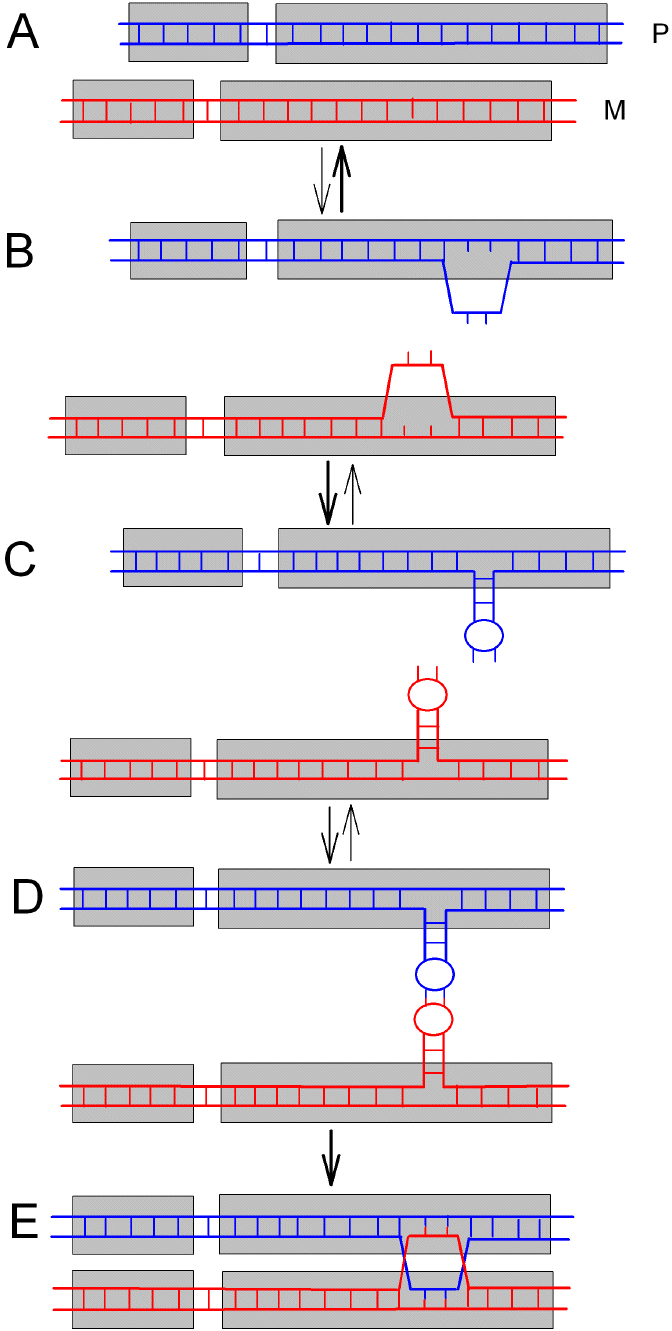 |
|
Figure 1.
Homology-recognition depends on prior formation of a stem-loop
intermediate. A. Two DNA duplexes of either paternal (P) origin
(in blue) or maternal (M) origin (in
red). The base pairing
between the two strands of each duplex is represented by the
laddering (short vertical lines). The two gray boxes indicate
sequence segments encoding two genes (microisochores) that
differ in base composition (GC%) values. B. Transient, unstable,
"bubble" intermediates with unpaired bases. For simplicity only
one strand is shown bubbled. The forms shown in B quickly return
to the forms shown in A (stable) or, if the sequences contain
palindromic segments, can transition to the stable stem-loop
conformations shown in C. Although stem-loop extrusions are
symmetric, affecting both strands equally, for simplicity only
one strand is stem-looped. The stem-loop conformations endure
for sufficient time to allow a "kissing" homology search by way
of the loops (D). If this succeeds (i.e. there is
complementarity between the bases in the loops), then a stable
intermediate is formed without strand breakage (E). The
cross-over points are known as Holliday junctions. |
Strand migration to the genic boundary
So whether recombination will occur between two
sequences depends on the extent to which their GC% values agree. What
has this to do with the Williams-Dawkins definition of the gene? The
crossover junctions within recombination complexes ("Holliday
junctions") can move along DNA to extend a region of paranemic pairing
(Figure 2). This branch migration is affected by the sequence (Sun et
al. 1998). Recent work suggests that migration proceeds when the
junction unfolds, and stalls when the junction folds - a process that is
sensitive to differences in GC% (Karymov et al. 2008). Thus, a shift in
GC% at the genic boundary should suffice to stop migration. Should the
initial cross-over have occurred within a gene, then the cross-over
point would proceed along a region of relatively uniform GC%
(corresponding to the gene) until it approached a region of different
GC% (intergenic DNA, neighbouring gene or intron). The change in GC%
would halt the migration. It is postulated that this would be sensed by
recombinogenic enzymes, which would then initiate strand-breakage. (For
genes with multiple exons, we should read exon instead of gene.) While
not excluding intragenic recombination, this process would greatly
decrease its probability. Thus the Williams-Dawkins gene would tend to
remain intact through the generations.
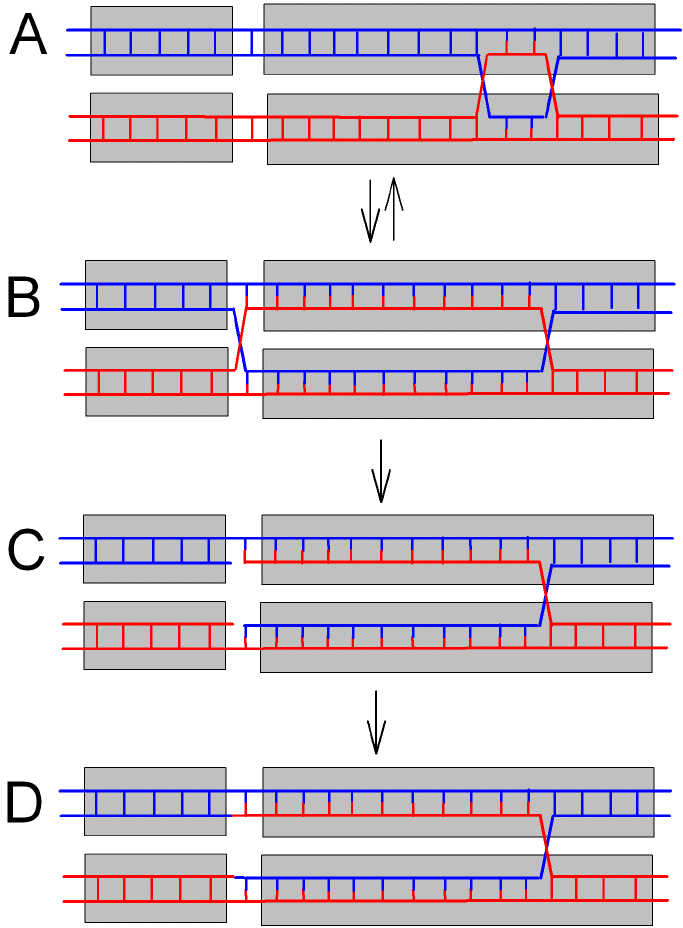 |
|
Figure 2 . Migration of a Holliday junction to the genic boundary where the change in base composition triggers strand breakage. A. The cross-over intermediate shown in Figure 1. B. Branch migration leftward to the genic boundary without strand breakage. C. Strand breakage. D. Ligation so that a maternal strand (red) is joined to a paternal strand (blue). The other Holliday junction can migrate to the right. The complex is resolved by mechanisms described in standard texts. |
Since there is a positive association between
palindromes (i.e. stem-loop potential) and recombinational crossing-over
(Inagaki et al. 2009), then also working to protect genes from
recombinational disruption would be the lower potential to extrude
stem-loops in exons than in introns and flanking DNA (Forsdyke 2006:
207-224). Indeed, fine-scale maps show that recombination crossover
points mainly occurs outside human genes (McVean et al. 2004; Coop et
al. 2008).
These considerations emphasize the positive role
of GC% identity in the successful pairing of allelic genes, but of equal
significance is the negative role of GC% non-identity in
preventing the pairing of
non-allelic genes, both somatically and in germ-line cells. This has
implications for the successful duplication both of genes and of genomes
(speciation). When GC% non-identity extends to entire meiotic
chromosomes then gametogenesis fails, a condition propitious for
divergence into two species. The case has been made from evidence, both
historic and modern, that reproductive isolation that leads to
speciation can initially be secured by virtue of differences in GC%
(Forsdyke 2010).
Historical antecedents of the selfish gene
concept date back to the nineteenth century. Williams and Dawkins went
further in defining the gene in terms of its ability to resist
recombinational disruption. But there need be no fundamental discrepancy
between their definition and the conventional definitions of
biochemists. Numerous studies, especially those of biophysicist Akiyoshi
Wada and colleagues (Wada et al. 1975; Wada and Suyama 1986), have shown
that, in addition to encoding a distinctive function dependent upon
primary sequence (base order), within the same boundaries each gene has
a distinctive, and to some extent
independent, base composition. The latter (GC%) is a gene's "accent"
that has the potential to determine whether paranemic pairing (no strand
breakage) between it and its allele can occur. The postulate that
recombinational strand breakage can proceed when GC% uniformity is lost
(i.e. at conventional genic boundaries) brings the two definitions into
close correspondence.
Acknowledgement
Queen's University hosts my web-pages which display some works of
Michael Guyer and several of the cited references including the
Third Report of the Commissioners
on the Cattle Plague (http://www.queensu.ca/academia/forsdyke/homepage).
This paper is dedicated to the memory of George Williams, who died on 7th
September 2010.
Baldwin GS, Brooks NJ, Robson RE, Wynveen A, Goldar A, Leikin S, Seddon
JM, Kornyshev AA (2008) DNA double helices recognize mutual sequence
homology in a protein free environment. Journal of Physical Chemistry B
112: 1060-1064.
Barzel A, Kupiec M (2008) Finding a match. How do homologous sequences
get together for recombination? Nature Reviews Genetics 9: 27-37.
Bateman JR, Wu C ting (2008) A genomewide survey argues that every
zygotic gene product is dispensable for the initiation of somatic
homolog pairing in Drosophila. Genetics
180: 1329-1342.
Bateson W (1904) Practical aspects of the new discoveries in heredity.
Memoirs of the Horticultural Society of
Bateson W (1919) Science and nationality.
Beale LS (1866) Letter to Charles Darwin. In:
The Correspondence of Charles Darwin (Burkhardt F, Porter DM, Dean
SA, Evans S, Innes S, Sclater A, Pearn A, White P, eds.), Vol. 14, pp
50-51.
Bibb MJ, Findlay PR, Johnson MW (1984)
The relationship between base composition and codon usage in bacterial
genes and its use for the simple and reliable identification of
protein-coding sequences.
Gene
30:
157-166.
Blumenstiel JP, Fu R, Theurkauf WE, Hawley RS (2008) Components of the
RNAi machinery that mediate long-distance chromosomal associations are
dispensable for meiotic and early somatic homolog pairing in
Drosophila melanogaster.
Genetics 180: 1355-1365.
Bultrini E, Pizzi E, Giudice PD, Frontali C (2003) Pentamer vocabularies
characterizing introns and intron-like intergenic tracts from
Caenorhabditis elegans and Drosophila melanogaster.
Gene 304:
183-192.
Bungener P, Buscaglia M (2003) Early connection between cytology and
Mendelism: Michael F. Guyer's contribution. History and Philosophy of
the Life Sciences 25: 27-50.
Chargaff E (1963) Essays on Nucleic Acids.
Cock AG,
Coop G, Wen X, Ober C, Pritchard JK, Przeworski M (2008) High-resolution
mapping of crossovers reveals extensive variation in fine-scale
recombination patterns among humans. Science 319: 1395-1398.
Crick F (1971) General model for the chromosomes of higher organisms.
Nature 234: 25-27.
Danilowicz C, Lee CH, Kim K, Hatch K, Coljee VW, Kleckner N, Prentiss M
(2009)
Single molecule detection of direct, homologous, DNA/DNA pairing.
Proceedings of the National
Dawkins R (1976) The Selfish Gene, pp. 25-38.
Doyle GG (1978) A general theory of chromosome pairing based on the
palindromic DNA model of Sobell with modifications and amplification.
Journal of Theoretical Biology
70: 171-184.
Edwards AWF (1998) Natural selection and the sex ratio: Fisher's
sources. American Naturalist 151: 564-569.
Eguchi Y, Itoh T, Tomizawa J-I (1991) Antisense RNA. Annual Review of
Biochemistry 60: 631-652.
Filipski J, Thiery JP, Bernardi G (1973) An analysis of the bovine
genome by Cs2SO4 Ag+ density
centrifugation. Journal of Molecular Biology 80: 177-197.
Forsdyke DR (1996) Different biological species "broadcast" their DNAs
at different (C+G)% "wavelengths."
Journal of Theoretical Biology
178: 405-417.
Forsdyke DR (2004) Regions of relative GC% uniformity are
recombinational isolators.
Journal of Biological Systems
12: 261-271.
Forsdyke DR (2007) Molecular sex: the importance of base composition
rather than homology when nucleic acids hybridize.
Journal of Theoretical Biology
249: 325-330.
Forsdyke DR (2009) Scherrer and Josts' symposium. The gene concept in
2008. Theory in Bioscience
128: 157-161.
Forsdyke DR (2010) George Romanes, William Bateson, and Darwin's "weak
point." Notes and Records of the Royal Society 64: 139-154.
Galton F (1894) Discontinuity in evolution. Mind 3: 362-372.
Grafen A, Ridley M (2007) Richard Dawkins: How a Scientist Changed the
Way We Think.
Grantham R (1980) Workings of the genetic code.
Trends in Biochemical Science
5: 327-331.
Grantham R, Perrin P, Mouchiroud D (1986) Patterns in codon usage of
different kinds of species.
Griffith F (1928) The significance of pneumococcal types. Journal of
Hygiene 27: 113-159.
Haldane JBS (1932) The Causes of Evolution, pp. 123-131.
Hamilton WD (1964) The genetical theory of social behaviour. Journal of
Theoretical Biology 7: 1-32.
Holliday R (1968) Genetic recombination in fungi. In:
Replication and Recombination of Genetic Material
(Peacock WJ, Brock RD, eds.), pp. 157-174. Camberra:
Hurst CC (1904) Mendel's discoveries in heredity. Transactions of the
Huxley TH (1893) Darwiniana. Collected Essays.
Inagaki
H, Ohye T, Kogo
H, Kato T, Bolor
H, Taniguchi M,
Shaikh TH, Emanuel BS,
Kurahashi H (2009)
Chromosomal instability
mediated by non-B DNA: Cruciform conformation and not DNA sequence is
responsible for recurrent translocation in humans.
Genome Research
19:
191-198.
Inoue S, Sugiyama S, Travers AA, Ohyama T (2007) Self-assembly of
double-stranded DNA molecules at nanomolar concentrations. Biochemistry
46: 164-171.
Johannsen W (1923) Some remarks about units of heredity. Hereditas 4:
133-141.
Karymov MA, Boganov A, Lyubchenko YL (2008) Single molecule fluorescence
analysis of branch migration of Holliday junctions: effect of DNA
sequence. Biophysical Journal 95: 1239-1247.
Kleckner N, Weiner BM (1993) Potential advantages of unstable
interactions for pairing of chromosomes in meiotic, somatic and
premeiotic cells.
Kornyshev AA (2010) Physics of DNA: unraveling hidden abilities encoded
in the structure of 'the most important molecule.' Physical Chemistry
Chemical Physics 12:
12352-12378.
Kornyshev AA, Wynveen A (2009) The homology recognition well as an
innate property of DNA structure. Proceedings of the National
Lao PJ,
Lobry JR, Sueoka N (2002) Asymmetric directional mutation pressures in
bacteria. Genome Biology 3
(10):research 0058.
McLuhan M (1964) Understanding Media:
The Extensions of
McVean GAT, Myers SR, Hunt S, Deloukas P, Bentley DR, Donnelly P (2004)
The fine-scale structure of recombination rate variation in the human
genome. Science 304: 581-584.
Moore G, Shaw P (2009) Improving the chances of finding the right
partner. Current Opinion in Genetics and Development
19: 99-104.
Morgan TH (1926) The Theory of the Gene.
Olby R (1974) The Path to the Double Helix.
O'Malley MA (2009) What did
Paz A, Kirzhner V, Nevo E, Korol A (2006) Coevolution of DNA-interacting
proteins and genome "dialect." Molecular Biology and Evolution 23:
56-64.
Roberts HF (1929) Plant Hybridization before Mendel. Princeton:
Romano TM (2002) Making Medicine Scientific. John Burdon Sanderson and
the Culture of Victorian Science, pp. 55-74.
Roux W (1881) Der Kampf der Theile im Organismus.
Schaap T (1971) Dual information in DNA and the evolution of the genetic
code.
Journal of Theoretical Biology
32: 293-298.
Sobell HM (1972) Molecular mechanism for genetic recombination.
Proceedings of the National
Simon J (1865) Minutes of Evidence Taken Before the Cattle Plague
Commissioners.
Spencer JP, Cecil RATG, Lowe R, Playfair L, Read CS, Jones HB, Quain R,
Parkes EA, Wormald T, Ceely R, Spooner C (1866) Third Report of the
Commissioners Appointed to Inquire into the Origin and Nature, etc. of
the Cattle Plague.
Stotz K (2009) Experimental philosophy of biology: notes from the field.
Studies in History and Philosophy of Science 40: 233-237.
Sueoka N (1999) Two aspects of DNA base composition: G+C content and
translation-coupled deviation from intra-strand rule of A = T and G = C.
Journal of Molecular Evolution 49: 49-62.
Sun W, Mao C, Liu F,
Vries H de (1889) Intracellulare Pangenesis.
Wada A, Suyama A (1986) Local stability of DNA and RNA secondary
structure and its relation to biological functions. Progress in
Biophysics and Molecular Biology 47: 113-157.
Wada A, Tachibana H, Gotoh O, Takanami M (1975) Long range homogeneity
of physical stability in double-stranded DNA. Nature 263: 439-440.
Wagner RE, Radman M (19750 A mechanism for
initiation of genetic recombination.
Proceedings of the National
Wang X, Zhang X, Mao C,
Williams GC (1966) Adaptation and Natural Selection. A Critique of Some
Current Evolutionary Thought, pp. 22-25.
Princeton:
Williams GC (1985) A defense of reductionism in evolutionary biology.
Williams GC (1992) Natural Selection: Domains, Levels, and Challenges,
pp. 10-13.
Wilson EB (1925) The Cell in Development and Heredity, 3rd ed., p. 928.
Wilson JH (1979) Nick-free formation of reciprocal heteroduplexes: a
simple solution to the topological problem.
Proceedings of the National
Zhang C, Xu S, Wei J-F, Forsdyke DR (2008) Microsatellites that violate Chargaff's second parity rule have base order-dependent asymmetries in the folding energies of complementary DNA strands and may not drive speciation. Journal of Theoretical Biology 254: 168-177.
![]()
| Journal | Date submitted | Identification code | Date rejected (accepted) |
| Biological Reviews | 15 April 2009 | BRV-04-2009-0044 | 11 May 2009 |
| Biology and Philosophy | 17 May 2009 | BIPH365 | 24 Aug 2009 |
| Journal of Theoretical Biology | 27 Aug 2009 | JTB-D-09-00685 | 3 Sept 2009 |
| Journal of Heredity | 14 Sept 2009 | JOH-2009-196 | 1 Dec 2009 |
| Journal of Genetics | 11 Jan 2010 | JGEN-D-10-00010 | 6 Apr 2010 |
| Notes & Records of the Royal Society | 5 Sept 2010 | RSNR-2010-0077 | 1 Nov 2010 |
| Biological Theory | 2 Nov 2010 | BIOT-D-10-00019 | 15 Dec 2010 (accepted) |
End Note on Hotspots (November 2011)
|
Although not extending to the above selfish gene scenario, geneticists approached the recombination problem through consideration of recombination hot spots, the location and DNA sequences of which were found to be highly species specific, and thus transient on an evolutionary time scale. So humans and chimpanzees did not have common hotspots. Some 25000 to 50000 hotspots were detected in the human genome by Myers et al. (2005), perhaps extending to 80000 (Khil & Camerini-Otero 2010). This number lay intriguingly close to the total number of exons. Using antibodies to a protein that localizes at putative recombination-initiated sites in mice, Smagulova et al. (2011) reported a central motif, flanked on the "top" strand by a purine-loaded segment to the left and a pyrimidine-loaded segment to the right. Thus, in extruded structures one would expect large purine-rich "kissing" loops to the left and large pyrimidine-rich "kissing" loops to the right (and the converse on the "bottom" strand). Hotspots were found mainly to localize to intergenic regions and introns (where the potential to form stem-loop structures is greater and Chargaff's second parity rule applies more strictly). As random mutations bring about changes in base sequence and composition (e.g. changes in GC%), the central canonical consensus motif would vary, and proteins interacting with that motif would be on a mutational "treadmill" (contrasting with proteins reacting with a more stable substrate, such as glucose; Paz et al. 2006; Forsdyke 2011). Thus, a most rapidly evolving positively-selected gene is that encoding the meiosis-specific "PR domain containing protein-9" (PRDM9), whose zinc finger segments bind to the DNA motif. Some 40% of currently identified human hotspots match the binding specificity of human PRDM9 (Ponting 2011). Mouse PRDM9 matches 73% of mouse hotspots (Smagulova et al 2011). Hailed as a "speciation gene" in vertebrates, the question arises as to whether the PRDM9 gene "tail" wags the genomic "dog", or vice-versa (Forsdyke 2011). Normally, within a species, identical motifs on homologous parental chromosomes would support strand pairing and meiotic recombination cross-over. However, since the motifs vary within a species, there is a chance of mismatch (i.e. heterozygosity). Recombination with gene conversion can follow, and the direction of the conversion is usually such as to eliminate the consensus in favour of the deviant. (Why this should be is another question.) However, although the consensus motif may disappear at a particular site, it will be reformed at other sites by mutations affecting cryptic motifs that differ only slightly from the consensus motif (Wahls and Davidson 2011). Thus, within the population, there will be a continuing flux of motifs around the consensus and, from time to time, individuals enriched in the same deviant motifs will meet and reproduce, hence tending to initiate a new line. When members of this line meet members of the consensus line, there will come a stage when the extent of the heterozygosity difference is such that recombination will fail. There will then be no gene conversion and a state of incipient reproductive isolation will exist (potential speciation). Forsdyke DR (2011) Evolutionary Bioinformatics, 2nd Edition. Springer, New York, pp. 238-239. Khil PP, Camerini-Otero RD (2010)Genetic crossovers are predicted accurately by the computed human recombination map. PLOS Genetics 6, e1000831. Myers S, Bottolo L, Freeman C, McVean G, Donnelly P (2005) A fine scale map of recombination rates and hotspots across the human genome. Science 310, 321-324. Ponting CP (2011) What are the genomic drivers of the rapid evolution of PRDM9? Trends in Genetics 27, 165-171. Smagulova F, Gregoretti IV, Brick K, Camerini-Otero RD, Petukhova GV (2011) Genome-wide analysis reveals novel molecular features of mouse recombination hotspots. Nature 472, 375-378. Wahls WP, Davidson MK (2011) DNA sequence-mediated, evolutionarily rapid, redistribution of meiotic recombination hotspots. Genetics 189, 685-694. |
End Note on Akiyoshi Wada (October 2014)
| Wada's pioneering studies mentioned above were but a small part of an illustrious career. As described by Cyranoski (2009 Nature 460, 171-172): "In the 1970s Wada ran into many sceptical biologists when he was one of the first to envision large scale automated genomic sequencing. But even as these technologies were ramping up elsewhere, Japan's bureaucrats stalled, its genomics fell behind, ... The unfolding of Wada's failed efforts are described in a book aptly titled A Defeat in the Genome Project." Much too late, in 1998 he became founding director of the RIKEN Genomic Sciences Center in Yokahama. One may speculate that, had the bureaucrats paid as much attention to Wada as to Kimura (the pioneer advocate of the "neutral theory" so loved of the biomathematicians, which is falling out of favour), the story might have been very different. |