The following paper was part of the
case I made in the 1990s that introns reflected the pressure for
stem-loop potential in genomes. Difficulties getting other papers in the
series (see elsewhere in these web-pages) accepted for publication, led to this one (drafted in
1995 and last accessed and saved in 1998) never being submitted for
publication. However, a decade later Jeffares, Penkett & Bahler
(2008
Trends in Genetics
24,
375-378) reported "that genes with rapidly changing expression
levels in response to stress contain significantly lower intron
densities" and proposed that introns were "selected against
in genes whose transcripts require rapid adjustment for survival of
environmental challenges." Although they invoked many good reasons why
this might be so, the stem-loop secondary structure of nucleic acids
was not mentioned. Accordingly, this paper was unearthed, dusted off, and placed here (with minimal further
editing; e.g. removal of some superfluous references). A more
exhaustive examination of heat-shock genes would seem necessary before
formal submission to a journal.
ADDED NOTE July 2015: FORS-D
analysis of nucleic acid structure has now been supplemented by other
approaches that have been applied to multiple sequences. In plant
seedlings (Arabidopsis thaliana) it is found that "mRNAs
associated with stress responses tend to have more single-strandedness,
longer maximal loop length and higher free energy per nucleotide
[i.e. less cohesive structure]" (Ding et al. 2014.
Nature 505,
696-700). Thus, they discovered that "genome-wide relationships
exist between in vivo mRNA structures and biological functions of the
encoded proteins." They suggest that "stress-response
RNAs may be more plastic, changing their structure in response to
changing cellular conditions." Indeed, the positive
FORS-D values reported below, indicate that base order has evolved to
actively prevent the formation of higher ordered
structure. |

Negative
base order-dependent stem-loop potential of heat shock protein 70 genes
indicates evolutionary selection to avoid mRNA secondary structure
D.
R. Forsdyke
[ draft unsubmitted paper]
Keywords: recombination, stem-loop, homology
search, introns, heat-shock protein, (G+C)/(A+T) ratio, speciation.
Abbreviations: FONS = folding of natural seqence;
FORS-M = folding of randomized sequence mean; FORS-D = folding of randomized
sequence difference.
Running head: Stem-loop Model for Initiation of
Recombination
Abstract
There has been an evolutionary selection
pressure on
base order favouring
the distribution of stem-loop potential throughout genomes.
If this pressure can be accommodated by the use of synonymous codons
and conservative amino acid exchanges, then long coding regions are possible.
Failing this, proteins are encoded in segments of low stem-loop potential (exons)
interrupted by regions of high stem-loop potential (introns). This is
particularly evident in genes under strong positive Darwinian selection. The
intronless heat shock protein 70 gene turns out to be a special case. Being
highly conserved (i.e. under negative evolutionary selection pressure), and
with one of the longest know open reading frames, high stem-loop potential
would be expected. However, the opposite is found.
It is suggested that heat shock protein 70 proteins must be synthesized
rapidly in response to intracellular stresses. This requires rapid
transcription under circumstances which might impair RNA splicing, and
minimization of RNA secondary structure to permit rapid translation. The
potential to form stem-loops appears to have been decreased by these needs.
Introduction
Heat-shock
70 proteins are synthesized rapidly in response to various biological,
chemical or physically stresses, such as viral infection, ethanol and
heat-shock. The genes are present in all cellular organisms and the sequences
are highly conserved. A role in intracellular self/not-self discrimination has
been suggested.
Heat-Shock Protein 70 as a Special Case
Among intronless genes are those encoding the heat shock proteins 70,
which are very highly conserved between species. The single open reading frame
of a human gene (HUMHSP70D) encodes a protein of 641 amino acids,
corresponding to a sequence of 1923 nt. This is one of the longest
uninterrupted open-reading frames known (Hawkins, 1988). Heat shock proteins
usually need to be synthesized rapidly in response to various intracellular
stresses (Forsdyke, 1994), and the absence of introns in the corresponding
gene implies a need to minimize delay in transcript processing (which might
itself be impaired by the heat-shock; Yost and Lindquist, 1986). The evolution
of the sequence could have served this need.
|
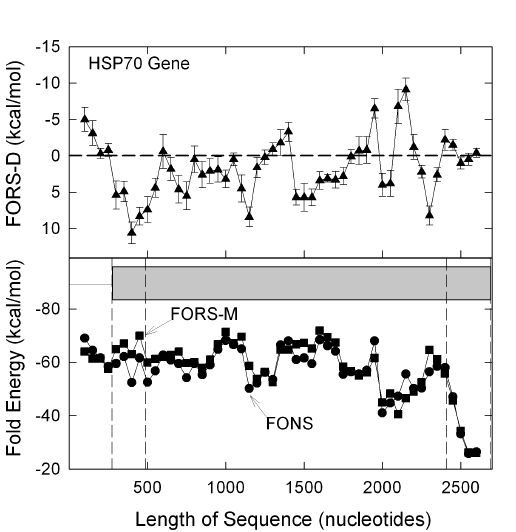
|
| FIG. 1. Fold energy minimization values (FORS-M,
FONS) and differences (FORS-D) for the 2691 nt sequence corresponding to a human heat shock protein
70 gene, HUMHSP70D. There are 51 windows of 200 nt, beginning at 50 nt
intervals (thus each overlaps its preceding neighbour by 150 nt). The
beginning of the mRNA corresponds to a window from nt 274 to nt 473. The last
window spans nt 2474-2673. The grey box indicates the exon. Vertical dashed lines in
the lower figure indicate, from left to right, the beginning
of the exon, the beginning of the protein-encoding region, the end of the
protein coding region, and the end the exon.
[Original plot prepared 27-3-1994 from file HUMHSP70D contributed to
GenBank 31-7-1992 by Hunt & Morimoto (1985)
Proc.
Natl. Acad. Sci. USA
82,
6455.]
|
Figure 1 shows that, in contrast to many long open reading
frame-containing genes
[shown in other papers],
positive FORS-D values predominate in
HUMHSP70D. The average for the 39 datapoints corresponding to the coding
region is 1.69sd0.62 kcal/mol. Two other heat-shock protein 70 genes gave
similar results. Thus, a human gene located in the major histocompatibility
complex (HUMMHHSP) has an average FORS-D value in the coding region of
1.33sd0.57
kcal/mol. A mouse homolog (MUSHP7A2) has an average FORS-D value in the coding
region of 0.42sd0.91 kcal/mol. That this is not a general characteristic of
single exon genes was suggested by examining two histone genes. The average
FORS-D value in the coding region of a histone 3 gene (HUMHISPRM) is -5.30sd1.16
kcal/mol (9 datapoints). The corresponding value for a histone 4 gene
(HUMHIS4) is -2.08sd1.59 kcal/mol (6 datapoints).
Thus, heat shock protein 70 genes would seem to be special cases. An
evolutionary pressure for positive FORS-D values may have been countermanded
by the need for rapid synthesis of a precise protein structure for which there
are not appropriate redundant codons. Consistent with this, codon preference
analysis (Gribskov and Devereux, 1991) shows minimal usage of rare human
codons in HUMHSP70D (data not shown). Under heat shock conditions the use of
codons corresponding to abundant tRNAs might facilitate protein synthesis.
Similarly, ribosomes might transverse most rapidly mRNAs transcribed from
genes which had evolved to decrease DNA (and hence RNA) secondary structure.
Acknowledgements
This work was supported by a grant from the Medical Research Council of
Canada.
LITERATURE CITED
FORSDYKE, D. R. 1994. The heat shock response
and the molecular basis of genetic dominance. J. Theor. Biol. 167:1-5.
FORSDYKE, D. R. 1995a. Fine-tuning of
intracellular protein concentrations, a collective protein function involved in aneuploid lethality,
sex-determination and speciation? J. Theor. Biol. 172:335-345.
FORSDYKE, D. R. 1995b. Relative roles of primary
sequence and (G+C)% in determining the hierarchy of frequencies of complementary trinucleotide
pairs in DNAs of different species. J. Mol. Evol. (submitted)
FORSDYKE, D. R. 1995c. Selective pressures on
DNA generate antisense phenomena as by-products. J. Mol. Evol. (submitted)
FORSDYKE, D. R. 1995d. Different biological
species "broadcast" their DNAs at different
(G+C)% "wavelengths". J. Theor. Biol. (submitted)
FORSDYKE, D. R. 1995e. Conservation of stem-loop
potential in introns of snake venom phospholipase A2 genes. An application of FORS-D
analysis. Mol. Biol. Evol. (submitted)
FORSDYKE, D. R. 1995f. Paradoxical relationship
between stem-loop potential and substitution density indicates that retroviral quasispecies conserve
recombination
function more than protein function. J. Mol. Biol. (submitted)
GRIBSKOV, M., and J. DEVEREUX. 1991. Sequence
analysis primer. Stockton Press, New York.
HAWKINS, J. D. 1988. A survey of intron and exon
lengths. Nucleic Acids Res. 16:9893-9905.
HUYNEN, M. A., D. A. M. KONINGS, and P. HOGEWEG.
1992. Equal G and C contents in histone genes indicate selection pressure on mRNA secondary structure. J. Mol. Evol. 34:280-291.
YOST, H. J., and S. LINDQUIST. 1986. RNA
splicing is interrupted by heat shock and is rescued by heat shock protein synthesis. Cell 45,
185-193.

Return to: Introns papers
(Click Here)
Go to: Bioinformatics Index
(Click Here)
Go to: Homepage (Click Here)

Placed
here 2 August 2008 and last edited
11 Nov 2020
by Donald Forsdyke
![]()
