|
DISCOVERY
NOTE Genome
Analysis
End Note 2006
ABSTRACT
In 1993 Prahbu began a paper entitled "Symmetry observations in long nucleotide sequences" with the concise and elegant statement:
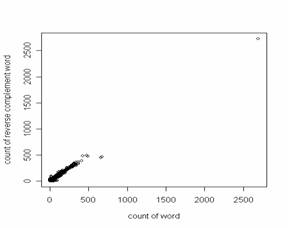
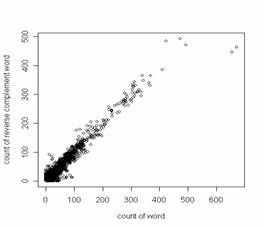
Thus, occurrences of 32 of the 64 trinucleotides (3-tuples) closely approximate to those of their 32 reverse complements (e.g. the frequency of TCA equals that of TGA), and occurrences of 128 of the 256 tetranucleotides (4-tuples) closely approximate to those of their 128 reverse complements (e.g. the frequency of TCAG equals that CTGA). Since Prabhu examined sequences of twenty-two species from a wide range of taxa, the symmetry principle, a manifestation of Chargaff's second parity rule (Forsdyke and Mortimer, 2000; Forsdyke, 2002; Forsdyke and Bell, 2004), appeared broadly applicable. Prabhu plotted the frequencies of oligonucleotides of a given length (n) against the frequencies of their corresponding complements. Since the frequencies were similar, the plots were rectilinear with a slope of 1.0 and intersected the origin. The correlation coefficient provided a measure of the extent to which the DNA of a particular species followed the principle. Baisnee et al., (2002) confirmed and extended Prabhu's observations using an "S1" metric ranging from zero (asymmetry) to one (symmetry). Prabhu was careful to note that the symmetry principle "extends to sets of n-tuples of higher order n with increase in length of the sequence." Higher n-tuples being rarer, although a sequence might contain several copies of a long oligonucleotide (e.g. a 12-tuple), it would be unlikely to contain an equal number of copies of the corresponding reverse complementary oligonucleotide (i.e. the symmetry would be less and the variance greater than in the case of smaller n-tuples). Thus, Baisnee et al., (2002) noted that "long sequences are necessary to accurately measure high order statistics and the corresponding symmetry," and showed that, for a given genome sequence, S1 decreases monotonically with oligonucleotide sequence length. For human chromosome 22 (approx. 33 Mb of sequence) they showed S1 = 1.00 for small n-tuples (n = 1-5), decreasing to 0.96 for 9-tuples. Using the same metric, Chen and Zhao (2005) extended this to the complete mouse, rat and human genomes from which they extracted sequences in 5 Mb windows. Symmetry measured as S1 decreased progressively with n-tuple length, arriving at a value of 0.4 for 12-tuples (i.e. in most segments 12-tuple occurrences were more asymmetric than symmetric with respect to occurrences of their reverse complements). PALINDROMIC
VERseS
"All DNA base sequences, regardless of their
origins or functions (coding versus non-coding) are messages written in
palindromic verses." Considering inverted repeats as
palindromic, these words of Susumu Ohno (1991) provided a proximate
explanation for the symmetry principle. The existence of closely located
inverted repeats implies that a DNA sequence has the potential to depart
from its classical duplex form and extrude stem-loops, a process that
would be facilitated by negative supercoiling (Murchie et al., 1992). The stems would contain n-tuples in one strand and
their reverse complementary n-tuples in the other strand. Of course,
symmetry alone is insufficient to ensure this. An n-tuple usually has to
be closely located to its complementary n-tuple within a strand in order
to become a stem in a stem-loop structure. The Nussinov-Forsdyke
hypothesis is that stem-loop potential has an adaptive advantage and
this has been an important factor driving the observed compositional
symmetry between n-tuples and their reverse complements (Nussinov, 1984;
Forsdyke, 1995a). This driving force has determined that complementary
n-tuple pairs be appropriately located. Using an energy-minimization approach, computer programs are able to fold single stranded nucleic acids into elaborate stem-loop structures, where the stems are formed by the stacking of Watson-Crick-paired bases. Thus, the local stem-loop potential of an RNA transcript, or of one strand of a DNA duplex, can be determined. Calculated stem n-tuples usually contain about 4 bases, but seldom as many as 12 bases (Fontana et al., 1993). Consistent with this, as noted above the S1 symmetry value for 12-tuples is only 0.4, indicating a decreased probability of a 12-tuple being able to interact with a reverse complementary 12-tuple to generate a stem.
Stem-loop
potential, greater than that of the corresponding shuffled sequences
(where the natural base order has been eliminated), is abundant and
widely distributed throughout genomes (Forsdyke, 1995a,b; seffens and
Digby, 1999; Cohen and Skiena, 2003). The fact that the potential is
greater in sequences that retain the natural base order implies that
stem-loop potential depends partly on primary sequence, and thus might
conflict with other DNA functions that depend on primary sequence.
Indeed, stem-loop potential is greater in introns than in exons where
the primary sequence encodes proteins (Forsdyke, 1995b,c), and where the
pressure to purine-load forces a departure from Chargaff's second
parity rule (Lao and Forsdyke, 2000; Forsdyke and Mortimer, 2000;
Forsdyke, 2001; Paz et al.,
2004; Forsdyke, 2006). Consistent with this, the symmetry principle
applies more strictly to introns than exons (Bultrini et
al., 2003). In 1984 Nussinov suggested that the symmetry might relate to "advantageous DNA structure." This is consistent with the idea that, for the homology search preceding DNA recombination, duplex strands must unpair ("unpairing hypothesis;" Crick, 1971) and extrude stem-loops (Sobell, 1972; Doyle, 1978; Forsdyke, 1995a-d). A relationship between stem-loop structures and recombination has long been known (reviewed in Lobachev et al., 1998). Indeed, recently Zhang et al. (2005) have shown that recombination break-points correspond to regions where the base order-dependent component of stem-loop potential is high. In this light it would be predicted that recombination rates would be optimized in sequence segments where there is close symmetry between n-tuples, thus facilitating stem formation (e.g. S1 = 1.00), and would either be unaffected, or inhibited (for reasons given below), when symmetry was impaired (e.g S1 < 1.00). However, the overall recombination rate within a DNA segment should be influenced by all its n-tuples, especially those that are abundant, those that are at frequencies close to those of their complementary tuples (i.e. exhibited symmetry), and those that are of lengths usually found in calculated structures (i.e. 4-tuples). A further wrinkle is that some n-tuples overlap, or are contained in, others. Finally, there is the problem that recombination rates exhibit different behaviors over large (Mb) and small (Kb) scales (Myers et al. 2005). In their paper Chen and Zhao (2005), without offering any justification, take one complementary n-tuple pair at a time. They examine how, irrespective off all the other n-tuple pairs in a 5 Mb segment, the symmetry in frequencies of an individual n-tuple and its complementary n-tuple is related to the recombination rate in that segment. They conclude: "Forsdyke suggested that because the stem-loop structure in supercoiled DNA facilitates the initiation of recombination, there is evolutionary pressure to produce reverse complement DNA sequences. If the local stem-loop structure is the only force for the reverse complement symmetry, the higher local symmetry levels should result in higher recombination rates. On the contrary, our analysis shows that there is a negative instead of a positive correlation between the local symmetry levels and the local recombination rates." Figure 2 of their paper is of particular interest. For the genomes of mice, rats and humans there is a negative correlation between recombination rates and symmetries, but only at high n-tuple levels (n = 10, 11 and 12). At these levels the 95% confidence intervals do not overlap zero correlation, indicating high significance. However, for the genomes of mice and rats there are positive correlations between recombination rates and symmetries at low n-tuple levels (n = 1-7). Here the 95% confidence intervals do, to varying degrees, overlap zero correlation, but since five values (mice) and seven values (rats) are all positive, the positive correlations must also be significant. In other words, for low n-tuples, despite the low variance between DNA segments in n-tuple symmetries, segments with marginally higher n-tuple symmetries have marginally higher recombination rates. For the human genome Chen and Zhao find very close to zero correlation at low n-tuple levels (n = 1-5), so here there is neither a positive nor a negative correlation. This, they regard as "not surprising because of the small variance of the calculated symmetry measures across different regions" at this n-tuple level. Thus, with the exception of a "not surprising" absence of positive correlation at low n-tuple values in the case of the human genome, the observations of Chen and Zhao do not negate the predicted positive correlation of compositional symmetry and recombination. Throughout their paper the authors focus on DNA regions that they acknowledge (see Figure 1 of their paper) display 12-tuple asymmetry. Yet they conclude that there is a "negative correlation between compositional symmetries and local recombination rates" (my italics). This statement is supported only at the long n-tuple level (see Figures 3-5 of their paper), where there is more variance between segments in degrees of symmetry so that determinations of correlations with recombination rates are facilitated.
At the RNA level, long stems have the potential falsely to activate intracellular alarms (reviewed in Forsdyke et al., 2002). Thus, there may have been some degree of negative selection of organisms with closely approximated pairs of long complementary n-tuples in transcribable nucleic acid segments. Such transcribable domains are likely to extend far beyond currently recognized genes (i.e. there is a "hidden transcriptome;" Forsdyke et al., 2002; Ota et al., 2004; Forsdyke, 2006). It would be anticipated that, to prevent translocation of a reverse complementary 12-tuple to the region of its forward complement (thus facilitating their co-transcription to generate an RNA species containing a long stem), recombination would be inhibited (perhaps by DNA sequence modifications that militate against stem-loop formation; Lang, 2005). In other words, organisms that had not, by chance, accepted mutations that impair recombination in the region, would have been negatively selected. Consistent with this, recombination rates decrease in DNA segments as the compromised symmetry between paired 12-tuples (S1 values around 0.4) becomes less compromised (i.e. symmetry increases; see Figures 3-5 of Chen and Zhao, 2005). An
argument against this explanation of the negative correlation is that
activation of intracellular alarms appears to require greater than
12-tuple stem lengths (i.e. at least two helical turns; n > 20).
However, it is possible that the 12-tuples which Chen and Zhao
identified had formed part of longer (perhaps imperfect) palindromes
that sufficed to activate alarms.
Achaz, G. et al. (2003) Associations between inverted repeats and the structural evolution of bacterial genomes. Genetics 164, 1279-1289. Baisnee, P.-F. et al. (2002) Why are complementary strands symmetric? Bioinformatics, 18, 1021-1033. Bultrini, E. et al. (2003) Pentamer vocabularies characterizing introns and intron-like intergenic tracts from Caenorhabditis elegans and Drosophila melanogaster. Gene, 304, 183-192. Chen, L. and Zhao, H. (2005) Negative correlation between compositional symmetries and local recombination rates. Bioinformatics, 21, 3951-3958. Cohen, B.
and Skiena, S. (2003) Natural selection and algorithmic design of mRNA. J.
Comput. Biol., 10,
419-432. Forsdyke, D. R. (1995a) Relative roles of primary sequence and (G+C)% in determining the hierarchy of frequencies of complementary trinucleotide pairs in DNAs of different species. J. Mol. Evol., 41, 573-581. Forsdyke,
D. R. (1995c) Conservation of stem-loop potential in introns of snake
venom phospholipase A2 genes. Mol. Biol. Evol., 12,
1157-1165. Forsdyke, D. R. and Mortimer, J. R. (2000)
Chargaff's legacy. Gene 261, 127-137. Murchie, A. I. H. et al. (1992) Helix opening transitions in supercoiled DNA. Biochim. Biophys. Acta, 1131, 1-15. Myers, S. et al. (2005) A fine-scale map of recombination hotspots across the human genome. Science 310, 321-324. Nussinov, R. (1984) Strong doublet preferences in nucleotide sequences and DNA geometry. J. Mol. Evol., 20, 111-119. Ohno, S.
(1991) The grammatical rules of DNA language: messages in palindromic
verses. In: Evolution of Life, pp. 97-108. Edited by Ota, T. et al. (2004) Complete sequencing and characterization of 21243 full-length human cDNAs. Nat. Genet., 36, 40-45. Paz, A. et al. (2004) Adaptive role of increased frequency of polypurine tracts in mRNA sequences of thermophilic prokaryotes. Proc. Natl. Acad. Sci. USA, 101, 2951-2956. Prabhu, V. V. (1993) Symmetry observations in long nucleotide sequences. Nucleic Acids Res., 21, 2797-2800. seffens, W. and Digby, D. (1999) mRNAs have greater negative folding free energies than shuffled or codon-choice randomized sequences. Nucleic Acids Res. 27, 1578-1584. Sobell, H. M. (1972). Molecular mechanism for genetic recombination. Proc. Natl. Acad. Sci. USA, 69, 2483-2487. Zhang, C.-Y. et al. (2005) The key role for local base order in the generation of multiple forms of China HIV-1 B'/C intersubtype recombinants. BMC Evol. Biol., 5, 53.
Journal peer review involves consulting with some of the peers judged to have expertise in the field under consideration. From among such peers (say A-E in no particular rank order) the Editors may pick two (say A and B), who may approve the paper. On publication the paper may then be seen by C-E, who may disapprove of the Editorial decision. This disapproval may take the form of a formal "letter" or, as in the case of the journal Bioinformatics, a "Discovery Note." In 2005 Bioinformatics published a paper entitled "Negative correlation between compositional symmetries and local recombination rates." The authors, Liang Chen and Hongyu Zhao specified, both in their Introduction and Discussion, that their results conflicted with those of one of the peers in their field (let us say E). In this circumstance, under normal circumstances Editors should, as a matter of good editorial practice, have included E among the peers initially consulted. However, this was not done. Accordingly E (the author of these web-pages) on detecting flaws in the paper, submitted a "Discovery Note." The Editors sent the note out for review (presumably to C and D) and, on the basis of arguments that E did not find convincing, declined to publish the correction. Thus, E, who did not know Chen and Zhao personally, has here published the paper on his web-pages (in May 2006), and has drawn the fact to the corresponding author's attention. Donald Forsdyke 26th May 2006 |