Thinking about Stem-Loops
![]()
Letter from Francis Crick to G. Khorana 28th June 1974 |
![]()
![]()
An Alternative Way of Thinking about Stem-Loops in DNA.
A Case Study of the Human G0S2 Gene
D. R. FORSDYKE
Journal of Theoretical Biology (1998) 192, 489-504 [With the permission of the copyright holder, Academic Press]
Relative Roles of Base Composition and Base Order
Randomization to Assess the Role of Base Composition
Importance of the Product of C and G
Single Linear Regression Correlations
Correlations with Other Base Composition-Derived Parameters
|
Summary. Single strands extruded from duplex DNA have the potential to form stem-loop structures, which may be involved in the homology search preceding recombination. The total stem-loop potential in a sequence window can be analysed in terms of the relative contributions of base composition and base order. There are at least ten base composition-determined parameters of relevance to the energetics of stem-loop formation. These are the quantities of the four bases themselves, and six derived parameters: ATmin, CGmin, Chargaff differences for the W and S bases, and two base products. The quantities of the least represented base of a Watson-Crick base pair (ATmin, CGmin) might provide an index of the total stem potential of a window. The degrees to which one base of a Watson-Crick pair exceeds the other (the Chargaff differences for the W bases and for the S bases) might provide an index of the total loop potential of a window. Base products (A x T, C x G) might provide an index both of stem and of loop potentials. Multiple regression analysis of the relationship of the ten parameters to the energetics of stem-loop formation in the G0S2 gene reveals major roles of S bases, and of base products. While base composition may primarily serve genome or genome sector "strategies", it becomes of local relevance in the case of CpG islands. Base order serves many local "strategies", whose demands may conflict. Base order serves the encoding of protein or of recognition motifs for regulatory factors. On the other hand there appear to be circumstances under which base order synergizes with, or antagonises, base composition in determining total stem-loop potential. Antagonism is evident when the base composition-dependent component of the stem-loop potential of a region is greater than the total stem-loop potential of that region. |
1. Introduction
Opportunities for the single strands of duplex DNA molecules to form stem-loop structures may arise both during replication, and when the duplex is supercoiled (Murchie et al., 1992). The former has been invoked to explain expansions and contractions of simple sequence repeats (Wells, 1996). The latter may be involved in the initiation of recombination by way of the "kissing" of the tips of single strand loops (Kleckner & Weiner, 1993; Kleckner, 1997). This self/not-self homology search would appear to be a fundamental part of various long-range intra- and inter-chromosomal homology-dependent phenomena (Lewis, 1954; Wu, 1993), and its progressive impairment could lead to speciation (Radman et al., 1993; Forsdyke, 1996a). The stem-loop potential of a DNA sequence is unlikely to be just a passive and indirect consequence of the action of various evolutionary pressures on DNA; there appear to be powerful genome-wide pressures which actively confer or inhibit the potential to form stem-loops (Forsdyke, 1995a,b).
Improved knowledge of the energetics of base stacking and loop formation has facilitated the development of computer programs to predict the most likely secondary structures (stem-loops) in nucleic acids using the energy-minimization approach (Zuker, 1994). The programs work well with single stranded RNA and, with minor adaptations, can be applied to single stranded DNA (Gacy et al., 1995; Nielsen et al., 1995). Although the chemistry and thermodynamics of stem-loop formation are very important, nucleic acids can be considered more simply as informational macromolecules with three fundamental characteristics: length, base composition and base order (Forsdyke, 1995c). If windows of constant length are chosen, the properties of stem-loop structures can be analysed just in terms of base composition and base order. The total potential of a window in a sequence to form stem-loop structures can be decomposed into the base composition-dependent stem-loop potential, and the base order-dependent stem-loop potential (Forsdyke, 1995d;1996b).
Studies using this approach led to the proposal that, because of a role in recombination, stem-loops once dominated the sequences of the "replicators" in the early "RNA world". Protein-encoding potential was subsequently imposed, and the pressure on base order to encode stem-loops had to accommodate to the pressure on base order to encode proteins (Forsdyke, 1995a). This suggested that a region of DNA might have a high base order-determined stem-loop potential if accommodation was possible, or zero base order-determined stem-loop potential if protein-encoding potential dominated. Computer analyses of the secondary structure of natural DNA sequences showed some agreement with this model, except that base order-determined stem-loop potential was often observed to be significantly less than zero. This was particularly evident in protein-coding regions, but was also found in non-protein-coding regions. I here extend the original model to deal with this, and demonstrate application of this way of thinking about stem-loops to the analysis of the contribution of various base composition-dependent parameters to the potential secondary structure of the human G0S2 gene. The results draw attention to the importance of base products, particularly CxG, and suggest a new approach to the detection of genes in the uncharted genomic sequences currently accumulating in databases.
2. Relative Roles of Base Composition and Base Order
The role of base order in determining the secondary structure of a nucleic acid is evident from the demonstrated importance of dinucleotide nearest-neighbour stacking interactions in calculating secondary structure (Borer et al., 1974; Breslauer et al., 1986). This may relate, in some way, to the observation that dinucleotide frequencies, rather than higher oligonucleotide frequencies, are a fundamental distinguishing feature of biological species (Nussinov, 1981; Forsdyke, 1995c). Calculation of secondary structure using the energy minimization approach requires accurate knowledge of the enthalpy and entropy changes involved in the formation of stems (which tend to stabilize secondary structure), and of various types of loops (which tend to destabilize the structure; Turner et al., 1988; Fontana et al., 1993; Sinden, 1994). The important process of assigning and refining energy values for formation of various structural elements is ongoing (Shen et al., 1995; SantaLucia et al., 1996), but there is growing confidence that the calculated secondary structures are biologically relevant, at least in the case of RNA (Zuker et al., 1991). The complexities in the calculations are ignored in the first part of the present work, and the energetics of nucleic acid folding are considered simply in terms of base composition and base order. Following Tinoco et al., (1971), "stability numbers" of 1 and 2 are assigned to A-T and C-G base pairs, respectively.
|
|
|
FIG. 1. Relative contributions of base order and base composition to the stability of stem-loops. Only bases in the stems are shown. The weakly bonded A-T base pairs (the W bases) are assigned an arbitrary score of one. The strongly bonded C-G base pairs (the S bases) are assigned an arbitrary score of two. Absence of base pairing scores as zero. The stability of a stem-loop is quantitated as the total stability score, which may be contributed to both by base order and by base composition. The terms at the bottom are described in the text. |
A series of hypothetical stem-loop structures are shown in Figure 1. For present purposes, only a single stem is allowed, and no sliding of strands relative to each other is permitted. Furthermore, the role of the loop is ignored. The first stem consists of a series of 16 A residues, which do not complement each other. Thus, if this were part of a natural DNA sequence, either the sequence would remain unextruded from duplex DNA, or a large loop (rather than a stem), would form. In the second stem a quarter of the bases are T residues.
|
If we assign the value 1 to an A-T base pair, then the stability of the stem can be scored as 4, with a high score meaning high stability. Since T residues are infrequent, it is likely that Ts will be opposite As, rather than opposite Ts (i.e. if As and Ts in the proportions 3:1 were allowed to randomly combine, there would be more A-Ts than T-Ts in the stem-loop). Thus base composition, rather than base order, can be considered to make the major contribution to the score. This is a fundamental point to which we will return. |
The third stem consists of equal proportions of As and Ts. The total possible number of A-T base pairs (8) is formed. The number of A-T pairs which would be formed if 8 As and 8 Ts were randomly mixed would be 4. Thus, the contribution to the score attributable to base composition alone is 4. By subtraction we can determine that the base order-dependent component of the score is 4. The fourth stem has the same base composition as stem 3, so that the potential base composition-dependent contribution to the stability remains at 4. However, the distribution of As and Ts is such that no A-T base pairs form. Thus, the total score is zero, and by subtraction we determine that the contribution of base order is minus 4. The bases are ordered so as to oppose the random tendency for a sequence of 8 As and 8 Ts to form 4 A-T intrastrand base pairs. Stem-loop extrusion from duplex DNA is opposed. Alternatively, if associated with an extruded structure, the order of bases would favour loop, rather than stem, formation.
The fifth and sixth stems contain equal proportions of all four bases. In the fifth stem only four complementary pairs are present. These are C-G pairs, to each of which we assign the score of 2, for a total score of 8. The relative contributions of base composition and base order are 3 and 5, respectively (as will be shown below). The sixth stem has no base pairing. The potential contribution of base composition remains at 3, so that the contribution of base order is minus 3.
Figure 1 shows that the correct bases may be present in the correct proportions, but if base order is inappropriate there may be no stem (zero total score). Stem stability depends on base composition in two ways.
Complementary bases must be present in equal proportions, and
the more C-G pairs there are, the higher will be the stability.
|
Since in biological sequences the first condition often applies (Prabhu, 1993; Forsdyke, 1995c), (C+G)% should be an important determinant of the contribution of base composition to stem-loop stability. If stem-loops are of critical importance for the initiation of recombination (Kleckner, 1997), then differences in (C+G)%, or derived parameters, should strongly influence the development and maintenance of recombination barriers (speciation). For example, viruses with similar genes (e.g. HIV-1 and HTLV-1), which are capable of co-existing in the same host cell, might destroy each other by mutually recombining (because of sequence similarities). Thus, each virus is part of the selective environment of the other. If (C+G)% differences could create a recombination barrier (while maintaining, through choice of appropriate codons, the abilities to encode similar proteins), then such differences would be selected for. Until each virus evolved a mechanism to prevent superinfection of its host cell by a second virus (analogous to prezygotic exclusion), there would be a sustained pressure for divergence of (C+G)% (Forsdyke, 1996a). |
3. Randomization to Assess the Role of Base Composition
If the fifth stem in Figure 1 were a natural sequence, then we could call the total score (8) the "folding of natural sequence" (FONS) value. How do we calculate the contributions of base composition and base order to that score? The stem has a unique characteristic, its base order, and two other characteristics which it shares with large sets of other possible DNA sequences, its length and base composition. The natural sequence is but one member of a hypothetical set of sequences which share length and base composition. Any average characteristic of this set must be a function of the latter two parameters. By keeping length constant, we can focus on the role of base composition. If the order of bases is randomized (shuffled), keeping the length constant, then members of the set which differ only in base order, are obtained. The sixth stem in Figure 1 is one member of the set. Figure 2 shows 10 other members of the set. These happened to be those generated first by consecutively shuffling base order in the fifth stem in Figure 1. Associated with each member is a total score. Each score is a "folding of randomized sequence" (FORS) value for the fifth stem in Figure 1.
|
|
|
FIG. 2. A set of stem-loop structures generated by successively randomizing (shuffling) the order of bases in the fifth stem in Figure 1. Total scores are determined as in Figure 1. |
It is seen that most members score less than the natural sequence. The mean score of the set of 10 is 3.3±0.7 (standard error), which closely approximates to 3. This is the "folding of randomized sequence mean" (FORS-M) value, which is an average characteristic of the set, and thus should be base order-independent. The value provides a measure of the contribution of base composition to the FONS value for the 5th stem of Figure 1. By subtraction, the contribution of base order is found to be 5. This is the "folding of randomized sequence difference" (FORS-D) value, which provides a measure of the contribution of the primary sequence (base order) to the stability of stem-loop structures in a natural sequence of given length. In this case, the FORS-D value is positive, and is significantly different from zero (P<0.01). This makes it likely that the sequence of the fifth stem in Figure 1 (if it were a real sequence) had accepted mutations which form complementary base pairs, thus enhancing its stem potential. FORS-D values can provide a measure of what Le & Maizel (1989) have termed "statistically significant" stem-loop potential. However, statistical significance may not be the same as functional significance (Karlin & Brendel, 1992). Le and coworkers (1991) have shown for retroviruses that "statistically significant" stem-loop potential is functionally relevant.
It will be noted that four of the stems derived by randomization (Fig. 2), have lower total scores than 3. An extreme example of this is the sixth stem in Figure 1. To generate by chance a stem with no base pairs would usually required more randomizations than the ten used to generate Figure 2. Thus the sixth stem of Figure 1 is relatively improbable. If the sixth stem were part of a real natural sequence, then the low FORS-D value would imply that base order had been working strongly against base composition in determining the total score. We return to this in section 9.
4. Importance of the Product of C and G
Of the various factors likely to contribute to the FORS-M value of a sequence window of a given length, the four simplest are the quantities of the four bases. Two slightly more complex factors are the bases of each potential base pair which are present in lowest amounts. Thus if the quantities of A, T, C and G in a 200 nt window are 60, 40, 70 and 30, respectively, then the "ATmin" would be 40 and the "CGmin" would be 30. These numbers would reflect the upper limit on the number of base pairs which could form stems, and might be expected to correlate positively with the base composition-dependent component of the stem-loop potential (FORS-M values).
Conversely, the excess of bases (in the example A - T = 20 and C - G = 40) might provide an indication of the minimum number of bases available to form loops. Since loops tend to destabilize stem-loop structures, these "Chargaff difference" values (Smithies et al., 1981; Bell & Forsdyke, 1999a, b), might be expected to correlate negatively with the base composition-dependent component of the stem-loop potential.
However, in section 2 above, FORS-M values were considered as if the result of random interactions between free bases in solution. This predicts that the two products of the quantities of pairing bases should be important (60 x 40, and 70 x 30, in the above example). The products would be maximal when pairing bases were in equal proportions (i.e. [50 x 50] > [60 x 40] > [70 x 30]). Thus the products provide an index both of the absolute quantities of the members of a Watson-Crick base pair, and of their relative proportions.
In an attempt to derive formulae permitting the prediction of FORS-M values directly from the proportions of the four bases, Chen and coworkers (1990) demonstrated the relative importance of eight of the above factors; (they referred to FORS-M values as "energyrandom" = "er" values). The factors were A, C, G, T, ATmin, CGmin, A x T, and C x G, where A, C, G, and T refer to the quantities of each particular base in a sequence window. Multiple least-squares linear regression analysis of data obtained by folding nucleic acid using the energy minimization approach (e.g. Zuker, 1994), produced coefficients for each of the factors which provided an index of relative importance. The products of the quantities of the Watson-Crick pairing bases (A x T, and C x G) were found to be of major importance, with the coefficients of C x G (the strongly interacting S bases), exceeding those of A x T (the weakly interacting W bases). Less important were ATmin and CGmin and the quantities of the four bases. The following case-study of the G0S2 gene supports these findings, and thus the line of reasoning presented in sections 2 and 3. The influence of the pairing of mismatched bases will not be considered here.
|
Comment: It is recommended that first time readers skip sections 5-7 and go straight to 8-10. (Click Here) |
In the simple model shown in Figures 1 and 2, high stem-loop potential was scored in positive stability units. However, in chemical thermodynamic terms, helix formation is strong to the extent that free energy is released. There is a loss of free energy, which is expressed in negative kilocalories/mol. The higher this negative value, the more stable the resulting structure. Figures 3a, b show the result of folding a real biological sequence using the energy minimization approach (Zuker, 1994). As in previous studies (Forsdyke, 1995a), ordinate scales are arranged such that values corresponding to stable stem-loop structures are uppermost. In the case of FONS and FORS-M values, high negative values (kcal/mol) are high on the ordinate scale (Fig. 3b). FORS-D values are derived by subtracting one of these from the other (Fig. 3a). The direction of the subtraction determines the sign of the FORS-D value. In earlier work this direction was such (FORS-M less FONS) that positive FORS-D values signified a high contribution of base order to stem-loop stability (Forsdyke, 1995a). In the present work the subtraction has been carried out the other way (FONS less FORS-M), so that negative FORS-D values now signify a high contribution to stem-loop stability. This makes it easier to relate FORS-M and FORS-D values.
|
|
|
FIG. 3. Fold analysis for 200 nt windows in the first 4400 nt of the sequence containing the human G0S2 gene (GenBank locus HUMG0S2PE). Fold energy values (FONS, FORS-M) and their differences (FORS-D ± standard errors of mean) were generated as described previously (Forsdyke, 1995a) using the energy minimization program LRNA (Zuker, 1994), and local data files with parameters for DNA (Nielsen et al., 1995). Base composition values for each window (open circles in (a)) were calculated as (C+G)/(W + S) and expressed as a percentage. Each data point corresponds to the middle of its 200 nt window. Each window overlaps the preceding window by 175 nt. The two exons are shown in (a) as large numbered boxes. From left to right, small boxes in the 5' flank refer to an AT-rich element showing strong dyad symmetry (stem-loop potential), an element with two sets of two repeats containing the sequence TCAGTTT, an element with two repeats each containing CCAAT sequences, and a region with 19 consecutive CT dinucleotides followed by 16 consecutive CA dinucleotides (Russell & Forsdyke, 1991). Vertical dashed lines indicate from left to right, the beginning of the CpG island (grey rectangle in (a)), the beginning of exon 1, the beginning of the protein-encoding region, the end of the protein-encoding region, and the end of exon 2. |
The gene analyzed is G0S2, a human CpG island-containing gene with two exons. The lectin-induced expression of the corresponding mRNA is highly sensitive to inhibition by cyclosporin A, consistent with involvement of the gene in the switch between the G0 and the G1 phases of the cell cycle (Russell & Forsdyke, 1991; Cristillo et al., 1997). The mouse homolog is involved in mesenchymal differentiation (reported by M. Ahrens, D. Schroeder & G. Gross in GenBank file MMG0S2). The 4400 nt human sequence serves present purposes well since the region of the exons is small, and there is extensive 5' flanking sequence to compare with this. Furthermore, the base composition, when expressed as percentage C+G, increases dramatically in the region of a CpG island (a region enriched in CpG dinucleotides; Cross & Bird, 1995; Fig. 3a). While base composition can often be considered as a genome or genomic sector "strategy", rather than a local "strategy" (Forsdyke, 1996a), in the region of the CpG island the high (C+G)% seems clearly a local phenomenon.
The base composition-determined component of the stem-loop potential (FORS-M) is relatively constant (about -25 kcal/mol) except in the region of the CpG island where the potential increases (to about -60 kcal/mol). The fluctuations of FONS values above the abscissa in the 5' flank (Fig. 3b) are observed to be largely base-order dependent, as reflected in the FORS-D values (high negative values; Fig. 3a). This is particularly apparent in the case of the AT-rich dyad (the most leftward of the open boxes).
If a major determinant of base composition-dependent stem-loop potential is the C x G product (see Section 4), then the high negative FORS-M values in the region of the CpG island could reflect the high (C+G)% of the region (Fig. 3a), but the relative proportions of C and G would have to be balanced. This implies a high CGmin. Conversely, the difference between the number of Cs and Gs ("Chargaff difference") should be low.
In the region of the CpG island, the pressure on base order to encode potential regulatory elements and protein might have countermanded the development of significant base order-dependent stem-loop potential. Thus, FORS-D values in this region might have fluctuated close to zero. However, as in previous studies with some other genes (Forsdyke, 1995b, d; 1996b), the values are consistently below the abscissa (i.e. positive values). This was not predicted in the stem-loop model as originally envisaged (Forsdyke, 1995a). The simple extension of the model set out in Figures 1 and 2 predicts that FORS-D values can descend significantly below the abscissa. FORS-D values below the abscissa are found in intergenic regions as well as within transcription units (Fig. 3a; Forsdyke, 1995a, b). Thus, values below the abscissa cannot easily be explained in terms of a need to avoid secondary structure in RNA (perhaps to facilitate translation). Base order-dependent stem-loop potential (FORS-D) appears to correlate negatively with base composition expressed as (C+G)%, whereas base composition-dependent stem-loop potential (FORS-M) appears to correlate positively (Figs. 3a, b). This will be further considered below.
6. Single Linear Regression Correlations
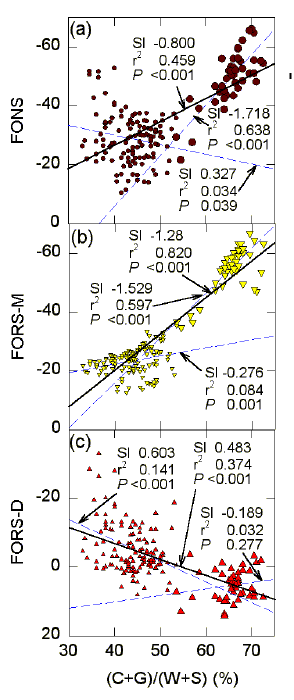 The significances of various correlations, some of
which are noted by direct inspection of Figure 3, were investigated using the
least-squares linear regression approach. Correlations with base composition expressed as
(C+G)% are shown in Figure 4. Points corresponding to the CpG island form
a cluster (larger symbols) at the right of the graphs. Linear regressions for all points (continuous dark lines) are
shown with linear regressions both for points corresponding to the CpG island, and for the
points remaining after excluding the CpG island (dashed blue lines). Some parameters of
the lines (3 sets of 3 numbers) are shown, with the uppermost set of numbers referring to
the linear regression for all points. The 3 numbers refer to the slope (Sl),
the coefficient of determination (r2), and the probability (P) that the slope
of the line is not significantly different from zero. The influence of base composition on
FONS values (Fig. 4a) reflects the summation of the dual influences on FORS-M values (Fig.
4b), and on FORS-D values (Fig. 4c).
The significances of various correlations, some of
which are noted by direct inspection of Figure 3, were investigated using the
least-squares linear regression approach. Correlations with base composition expressed as
(C+G)% are shown in Figure 4. Points corresponding to the CpG island form
a cluster (larger symbols) at the right of the graphs. Linear regressions for all points (continuous dark lines) are
shown with linear regressions both for points corresponding to the CpG island, and for the
points remaining after excluding the CpG island (dashed blue lines). Some parameters of
the lines (3 sets of 3 numbers) are shown, with the uppermost set of numbers referring to
the linear regression for all points. The 3 numbers refer to the slope (Sl),
the coefficient of determination (r2), and the probability (P) that the slope
of the line is not significantly different from zero. The influence of base composition on
FONS values (Fig. 4a) reflects the summation of the dual influences on FORS-M values (Fig.
4b), and on FORS-D values (Fig. 4c).
As the proportion of the S bases increases (and the proportion of the W bases reciprocally decreases), the base composition-dependent stem-loop potential increases (Fig. 4b). This emphasizes the positive role of the S bases. Points corresponding to the CpG island make a major contribution, and when these points are removed the dependence of FORS-M values on base composition is much less evident (r2 = 0.084), although still significant (P = 0.001). This emphasises the point that base composition tends to be a non-local, genome or genome-sector, characteristic, and tends to have a large local impact only under unusual circumstances (presence of a CpG island).
|
FIG. 4. Influence of base composition expressed as (C+G)% on (a) FONS, (b) FORS-M, and (c) FORS-D values (kcal/mol) for 200 nt sequence windows in the region of the human G0S2 gene. Points with large symbols corresponding to the CpG island (see Fig. 3). Least squares linear regression lines are either for all points (continuous lines and upper sets of three parameter values), or for the points corresponding to the CpG island (large symbols and dashed lines), or for points remaining after excluding the CpG island (small symbols and dashed lines). The sets of three values are from the standard Minitab output (Ryan & Joiner, 1994) and indicate the slope of the regression line (Sl), the square of correlation coefficient (r2), and the probability (P) that the slope of the line is not significantly different from zero. Probabilities that the slopes of the two dashed regression lines were not significantly different from each other were calculated using an interaction model with dummy qualitative variables (Ryan & Joiner, 1994). The resulting P values were (a) <0.001, (b) <0.001, (c) 0.003. For further details please see the legend to Figure 3. |
On the other hand, base order tends to be a local characteristic. Points for base order-dependent stem-loop potential are less clustered (vary more at the local level; Fig. 4c) than points for base composition-dependent stem-loop potential (Fig. 4b). Although, by definition, FORS-D values are what is left from the FONS values after the base composition-dependent component is subtracted, FORS-D values correlate negatively with base composition. As the proportion of the S bases increases, FORS-D values decrease (Fig. 4c), thus appearing to oppose the positive correlation of FORS-M values (Fig. 4b).
Although the slopes for the points corresponding to the CpG island, and for the points remaining after exclusion of the CpG island, are different (P=0.003), the two sets of points for FORS-D values (Fig. 4c) are not so clearly demarkated from each other as in the case of the two sets of points for FORS-M values (Fig. 4b). Thus FORS-D values more readily appear part of a continuum. FORS-M values for the CpG island indicate a discrete base composition-dependent perturbation of the sequence in the region of the CpG island to which base order-dependent potential (FORS-D values) has "responded" as part of a process which is evident even when values for (C+G)% are below 50 (i. e. the regression line for the non-CpG island points alone slopes down significantly).
As judged by FONS values (Fig. 4a), the FORS-D "response" is more than adequate when values for (C+G)% are below 50, and the slope of the linear regression plot for the points with the CpG island excluded (0.327), reflects a balance in favour of the FORS-D values (slope 0.603), rather than of the FORS-M values (slope -0.276). Thus, base order appears to "control" the total stem-loop potential (FONS values) in regions other than the CpG island, where base composition tends to dominate.
7. Correlations with Other Base Composition-Derived Parameters
The base composition of single stranded DNA, expressed as (C+G)% indicating (C+G)/100 bases, decreases as total A+T increases, and does not take into account the relative proportions of the bases of each Watson-Crick base pair. ATmin, CGmin, Chargaff differences, and the base products (A x T, C x G), provide some measure of changes in these proportions.
|
|
|
FIG. 5. Influence of ATmin, and CGmin, on (a, b) FONS, (c, d) FORS-M, and (e, f) FORS-D values (kcal/mol) for 200 nt sequence windows in the region of the G0S2 gene. ATmin and CGmin refers to the number of bases of the least represented member of a Watson-Crick base pair. The probabilities (P) that the slopes (dashed lines) are not significantly different from each other are (a) 0.003, (b) 0.012, (c) 0.018, (d) <0.001, (e) 0.092, (f) 0.981. For further details please see the legend to Figure 4. |
In Figure 5, points for the CpG island are again seen as a cluster (large symbols) corresponding to low ATmin values, and high CGmin values. Thus, in the region of the CpG island the quantities of the two S bases are high, and their relative proportions are similar (high CGmin), consistent with a contribution of stems to the high stem-loop potential values (Fig. 3). The slopes of the regression lines for all points (continuous lines) have different signs when plots for the W bases (Figs. 5a, c, e) are compared with corresponding plots for the S bases (Figs. 5b, d, f). Thus, base composition-dependent stem-loop potential (negative FORS-M values) decreases as ATmin increases, and increases as CGmin increases, whereas base order-dependent stem-loop potential (negative FORS-D values) increases as ATmin increases and decreases as CGmin increases. The base composition-dependent component dominates the folding of the natural sequence (FONS values increase as CGmin increases).
When points for the CpG island are removed from the analyses, the CGmin values continue strongly to affect the FORS-M values (slope -0.604; r2 = 0.794; P<0.001; Fig. 5d), but the base order-dependent stem-loop potential (FORS-D values) is independent of CGmin (Fig. 5f). Similarly, FORS-M values for points corresponding to the CpG island are strongly affected by the CGmin (slope -1.112; r2 = 0.687; P<0.001), but FORS-D values are independent of CGmin (slope 0.034; r2 =0.001; P=0.742). Only when all points are taken into account is there a dependence of FORS-D values on CGmin (Fig. 5f). When points for the CpG island are excluded, variations in ATmin do not affect FORS-M values (Fig. 5c), and have a barely significant affect on FORS-D values (Fig. 5e). FORS-M values for points corresponding to the CpG island are weakly influenced by variations in ATmin (P=0.025), whereas the corresponding FORS-D values are not influenced by variations in ATmin (P=0.290). These results emphasize the requirement of high and balanced levels of the S bases for the base composition-dependent component of the stem-loop potential in a sequence window, whereas the base order-dependent component becomes dependent on CGmin only when "normal" sequence is accompanied by a special region in which base composition has a local influence (CpG island; Fig. 5f).
|
|
|
FIG. 6. Influence of Chargaff differences for the W and S bases on (a, b) FONS, (c, d) FORS-M, and (e, f) FORS-D values (kcal/mol) for 200 nt sequence windows in the region of the G0S2 gene. Chargaff difference values for the W bases in each window (deltaW) were calculated as the absolute value of A-T (|A-T|). Chargaff difference values for the S bases in each window (deltaS) were calculated as the absolute value of C-G (|C-G|). The probabilities (P) that the slopes (dashed lines) are not significantly different from each other are (a) 0.192, (b) 0.086, (c) >0.1, (d) <0.001, (e) >0.1, (f) 0.186. For further details please see the legend to Figure 4. |
Figure 6 shows similar data for Chargaff differences. In the case of the W bases, Chargaff differences have no significant influence on the values specified by the two sets of points (large and small symbols; Figs. 6 a, c, e). On the other hand, for both sets of points the base composition-dependent stem-loop potential shows a significant decrease (less negative FORS-M values) as the number of potentially unpaired S bases (Chargaff difference for the S bases) increases (Fig. 6d). This again emphasises the major role of the S bases in determining FORS-M values, and indicates that the greater the number of unpaired S bases the greater would be the potential to form loop domains which would tend to destabilize stem-loop structures (make less negative the folding energy). When points for the CpG island (large symbols) are excluded, the base order-dependent stem-loop potential also shows a small decrease as Chargaff differences for the S bases increase (Fig. 6f). Essentially similar results are obtained when Chargaff differences are expressed relative to the total number of W or S bases in the same sequence window (i. e. dW/W, and dS/S; Bell & Forsdyke, 1999a,b).
|
|
|
FIG. 7 Influence of the products of the bases involved in classical Watson-Crick base pairing on (a, b) FONS, (c, d) FORS-M, and (e, f) FORS-D values (kcal/mol) for 200 nt sequence windows in the region of the G0S2 gene. The probabilities (P) that the slopes (dashed lines) are not significantly different from each other are (a) <0.001, (b) <0.001, (c) <0.001, (d)<0.001, (e) 0.036, (f) 0.005. For further details please see the legend to Figure 4. |
As expected (Section 4), the products of the Watson-Crick pairing bases correlate very well with FORS-M values (Fig. 7). This is seen best with the S bases (r2 = 0.924), and is still significant (P<0.001) when points corresponding to the CpG island are discarded (r2 = 0.378; Fig. 7d). Base order-dependent stem-loop potential (negative FORS-D values) decreases (values become less negative) as the products of the S bases increase (Fig. 7f), and this decrease is still evident when points corresponding to the CpG island are discarded. In the latter case, FORS-M and FORS-D values are in balance, so that FONS values for the S bases, show no significant correlation with the base product (Fig. 7b). When the CpG island is included a correlation is evident. In the case of the W bases, the results are the opposite of those for the S bases; increases in the W base product correlate with decreasing base composition-dependent stem-loop potential (FORS-M values became less negative), and with increasing base order-dependent stem-loop potential (FORS-D values became more negative).
In FORS-D plots, the slopes of the regression lines for the points corresponding to the CpG island (large symbols) are not significantly different from zero; the slopes of the two regression lines corresponding to the presence and absence of the points for the CpG island are very close (Figs. 7e, f); this indicates that the base order-dependent stem-loop potential acts in a uniform manner over a wide range of base product concentrations as part of a CpG island-independent continuum. However, the slopes (dashed lines) for the large and small symbols are different from each other at a low level of significance (P=0.36; P=0.005). This contrasts with the much higher levels of significance (P<0.001) for the differences between the slopes (dashed lines) for the FORS-M plots (Figs. 7c,d).
The multiple dependencies of FONS, FORS-M and FORS-D values shown individually in Figures 4-7 were further analyzed using the multiple regression approach (Ryan & Joiner, 1994). All data points were taken into account, including those for the CpG island. Chargaff difference values for the W and S bases, expressed as deltaW/W and deltaS/S, were added to the eight predictors employed by Chen et al., (1990; see Section 4). A first order linear model is designed to produce an intercept at the ordinate and a set of coefficients, one associated with each of the predictors. The best two results for increasing number of predictors, are shown in Table 1.
|
Potential predictor |
--> |
Individual bases |
Base pair products |
Minimum bases |
Chargaff differences |
Correlation coeff. stand. dev., & Cp |
||||||||
|
Fold measure |
Number of pre- dictors |
A |
T |
C |
G |
AxT |
CxG |
ATmin |
CGmin |
dW/W |
dS/S |
r2(%) |
S |
Cp |
|
FONS |
1 |
- |
- |
- |
- |
- |
- |
- |
+ |
- |
- |
65.4 |
7.426 |
70.3 |
|
" |
1 |
- |
- |
- |
- |
- |
+ |
- |
- |
- |
- |
58.1 |
8.170 |
119.8 |
|
" |
2 |
- |
- |
- |
- |
- |
+ |
- |
- |
- |
+ |
68.9 |
7.057 |
48.2 |
|
" |
2 |
- |
- |
- |
- |
+ |
+ |
- |
- |
- |
- |
66.5 |
7.328 |
64.8 |
|
" |
3 |
- |
- |
+ |
+ |
- |
+ |
- |
- |
- |
- |
76.0 |
6.212 |
1.7 |
|
" |
3 |
- |
- |
- |
- |
- |
+ |
- |
+ |
- |
+ |
71.8 |
6.744 |
30.8 |
|
" |
4 |
- |
- |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
76.5 |
6.147 |
0.7 |
|
" |
4 |
- |
- |
+ |
+ |
- |
+ |
+ |
- |
- |
- |
76.4 |
6.186 |
1.3 |
|
" |
5 |
- |
- |
+ |
+ |
+ |
+ |
- |
- |
+ |
- |
76.6 |
6.177 |
1.9 |
|
" |
5 |
- |
+ |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
76.6 |
6.179 |
2.0 |
|
" |
6 |
+ |
- |
+ |
+ |
+ |
+ |
- |
- |
+ |
- |
76.7 |
6.183 |
3.3 |
|
" |
6 |
- |
+ |
+ |
+ |
+ |
+ |
- |
- |
+ |
- |
76.7 |
6.186 |
3.4 |
|
" |
7 |
+ |
- |
+ |
+ |
+ |
+ |
+ |
- |
+ |
- |
76.7 |
6.200 |
5.2 |
|
" |
7 |
+ |
- |
+ |
+ |
+ |
+ |
- |
- |
+ |
+ |
76.7 |
6.201 |
5.2 |
|
FORS-M |
1 |
- |
- |
- |
- |
- |
+ |
- |
- |
- |
- |
92.4 |
4.178 |
1000 |
|
" |
1 |
- |
- |
- |
- |
- |
- |
- |
+ |
- |
- |
89.7 |
4.868 |
1417 |
|
" |
2 |
- |
- |
+ |
- |
- |
+ |
- |
- |
- |
- |
96.3 |
2.902 |
396 |
|
" |
2 |
- |
- |
- |
- |
- |
+ |
- |
- |
- |
+ |
96.0 |
3.046 |
453 |
|
" |
3 |
- |
- |
+ |
+ |
- |
+ |
- |
- |
- |
- |
98.2 |
2.039 |
113.1 |
|
" |
3 |
- |
- |
+ |
- |
+ |
+ |
- |
- |
- |
- |
97.4 |
2.463 |
239.0 |
|
" |
4 |
- |
- |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
98.4 |
1.939 |
87.5 |
|
" |
4 |
- |
- |
+ |
+ |
- |
+ |
+ |
- |
- |
- |
98.4 |
1.943 |
88.5 |
|
" |
5 |
- |
- |
+ |
+ |
- |
+ |
- |
+ |
- |
+ |
98.7 |
1.719 |
35.5 |
|
" |
5 |
- |
+ |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
98.7 |
1.776 |
48.6 |
|
" |
6 |
- |
+ |
+ |
+ |
- |
+ |
- |
+ |
- |
+ |
98.8 |
1.669 |
25.4 |
|
" |
6 |
- |
- |
+ |
+ |
- |
+ |
+ |
+ |
- |
+ |
98.8 |
1.669 |
25.4 |
|
" |
7 |
- |
+ |
+ |
+ |
- |
+ |
+ |
+ |
- |
+ |
99.0 |
1.575 |
6.6 |
|
" |
7 |
- |
+ |
+ |
+ |
- |
+ |
- |
+ |
+ |
+ |
98.9 |
1.580 |
7.6 |
|
FORS-D |
1 |
- |
- |
- |
- |
+ |
- |
- |
- |
- |
- |
35.7 |
6.763 |
32.8 |
|
" |
1 |
- |
- |
- |
- |
- |
+ |
- |
- |
- |
- |
34.3 |
6.837 |
37.2 |
|
" |
2 |
- |
- |
- |
- |
- |
- |
- |
+ |
- |
+ |
39.7 |
6.571 |
22.7 |
|
" |
2 |
- |
- |
- |
- |
- |
- |
+ |
- |
+ |
- |
39.5 |
6.582 |
23.3 |
|
" |
3 |
- |
- |
+ |
- |
- |
- |
- |
+ |
- |
+ |
43.4 |
6.381 |
13.0 |
|
" |
3 |
- |
- |
- |
+ |
- |
- |
- |
+ |
- |
+ |
42.6 |
6.428 |
15.6 |
|
" |
4 |
- |
- |
+ |
- |
- |
- |
- |
+ |
+ |
+ |
45.6 |
6.279 |
8.5 |
|
" |
4 |
- |
- |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
45.6 |
6.279 |
8.5 |
|
" |
5 |
- |
+ |
+ |
+ |
- |
+ |
- |
- |
+ |
- |
47.5 |
6.183 |
4.4 |
|
" |
5 |
- |
+ |
+ |
- |
- |
- |
+ |
- |
+ |
+ |
47.1 |
6.208 |
5.7 |
|
* A first order linear model for the prediction of FONS, FORS-M
and FORS-D values from various base-composition derived parameters was analysed using the
"best subsets regression" procedure (BREG) with Minitab software (Ryan &
Joiner, 1994). Folding energies (kcal/mol-1) and base compositions were
obtained from overlapping 200 nt sequence windows in the first 4.4 kb of the GenBank
sequence of the human G0S2 gene as shown in Figure 3. The best two results obtained
with each number of predictors are shown. Plus signs (+) indicate the most useful
predictors corresponding to a particular prediction level. † ‡ r2(%) is the square of the Pearson product moment correlation coefficient, expressed as a percentage, and not adjusted for degrees of freedom. § S is the standard deviation relative to the calculated least squares regression line, and expressed as kcal/mol. Cp is a statistic calculated as [(SSEp)/(MSEm)]-[n - 2p], where SSEp is the sum of squared errors obtained with the regression line corresponding to p predictors, MSEm is the mean squared error (variance) for the line corresponding to the full model which includes all 10 base composition-derived parameters, and n is the number of windows. If a model has little bias, Cp should be close to p. |
The best result is obtained using seven of the base composition-derived parameters to predict the base composition-dependent stem-loop potential (FORS-M values). The high correlation coefficient (r2 = 99%), and low standard deviation (1.575 kcal/mol), indicate that 99% of the variance can be accounted for in terms of the seven parameters. Only four parameters are required for the best prediction of the overall folding of the natural sequence (FONS values), but the standard deviation is high (6.174 kcal/mol) and the correlation coefficient (r2 = 76%) indicates that 23% of the variance in FONS values is accounted for by base order.
The base order-dependent stem-loop potential itself (FORS-D values) can be predicted, albeit with much variation, from five of the base composition-determined predictors (r2=47.5; s= 6.183 kcal/mol). Thus, although FORS-M values can be almost perfectly predicted from base composition-derived parameters, implying no contribution (direct or indirect) of base order, FORS-D values are partly determined by base composition-derived parameters, implying a response of base order to some aspect of the base composition of a sequence window.
The formulae corresponding to the optimum models of Table 1, are shown in Table 2. The S bases dominate, and the CxG product is of major importance. This supports the view that compatibility of (C+G)% is of fundamental importance for recombination; small deviations in (C+G)% should change the pattern of stem-loops thus impairing the "kissing" interactions required for the initiation of the homology search (Forsdyke, 1996a).
|
FONS = |
-103 |
. |
+1.59C |
+1.72G |
-0.0387[CxG] |
. |
. |
-0.0841[dW/W] |
|
-34.25 |
-103 |
. |
+79.5 |
+86.0 |
-96.75 |
. |
. |
-0 |
|
. |
||||||||
|
FORS-M= |
-37.4 |
+0.089T |
+1.19C |
+1.08G |
-0.0281[CxG] |
-0.136[ATmin] |
-0.74[CGmin] |
-0.386[dS/S] |
|
-33.50 |
-37.4 |
+4.45 |
+59.5 |
+54.0 |
-70.25 |
-6.8 |
-37 |
-0 |
|
. |
||||||||
|
FORS-D = |
-49.2 |
-0.182T |
+0.73C |
+0.938G |
-0.0124[CxG] |
. |
. |
-0.174[dW/W] |
|
-5.8 |
-49.2 |
-9.1 |
+36.6 |
+46.9 |
-31.0 |
. |
. |
-0 |
|
*Optimum models were derived using the best subsets regression procedure as described in Table 1. Numbers below the equations show values calculated for a hypothetical 200 nt window containing equal proportions of all four bases. |
9. Conflict between base order and base composition
Since base composition appears to be primarily a genome or genome sector "strategy", in the original model base composition-dependent stem-loop potential was considered of little local relevance (Forsdyke, 1995a, b). In coding regions the degeneracy of the genetic code would usually have allowed accommodation of coding information to base composition (Nichols et al., 1981; Kagawa et al., 1984). It was also possible that in non-coding regions, nucleic acid motifs recognized by regulatory proteins might show some degeneracy, permitting some accommodation to base composition. Thus, attention was focused on base order-dependent stem-loop potential as the primary parameter of relevance to local nucleic acid function.
|
|
FIG. 8. FORS-D values (D) support or oppose FORS-M values (M) under different conditions, to generate FONS values (N). (a) and (b) illustrate the original model (Forsdyke, 1995a), which postulated an evolutionary conflict just between protein pressure and base order-dependent stem-loop potential (FORS-D pressure). Base composition-dependent stem-loop potential (FORS-M pressure) was held to reflect a "global" genomic force of little local relevance. (a) represents a typical non-coding region where FORS-D supplements FORS-M. (b) represents a coding region where protein pressure has overwhelmed FORS-D pressure (i. e. FORS-D = 0). (c) and (d) extend the model by supposing some responsiveness of FORS-M values to local pressures, which may be accompanied by synergistic (c) or antagonistic (d) changes in FORS-D values. For further explanation please see the text.
The most usual situation is where FORS-M and FORS-D values simply summate to generate FONS values (Fig. 8a). Most genomes studied have average FORS-D values around -4 kcal/mol (Forsdyke, 1995c), which adds on to a generally much larger negative FORS-M value, to generate an even larger total stem-loop potential (negative FONS value). The initial model stated that FORS-D could be countermanded by other local pressures on base order so that in certain regions (e.g. exons) only the FORS-M value might contribute to the total stem-loop potential (Fig. 8b).
Around nucleotide 900 of G0S2 the base composition-determined component of the stem-loop potential (FORS-M values) falls "below" -25 kcal/mol (becomes less negative). In this region the base order-dependent component (FORS-D values) is positive (below the abscissa in Figure 3a), so that FONS has a very low negative value (Fig. 3b). In this case, FORS-M and FORS-D appear to synergize to "decrease" the observed FONS values (i.e. make them less negative). This is illustrated in Fig. 8c and indicates the existence of an evolutionary selection pressure on base order either to maintain DNA in duplex form (prevent stem-loop extrusion), or to maintain a very large loop if a stem could form. There is a very large Chargaff difference for the S bases around nucleotide 900 (C>G), indicating that in this case such loops, if they occur, would be very C-rich.
In the region of the CpG island FORS-M values are exceptionally negative (Figs. 3b, 4b). It appears that base composition-dependent stem-loop potential can respond to a local "strategy" (the island). Degeneracy of the code (or interchange of amino acids with similar function), and possible degeneracy of functional regulatory motifs, would be insufficient to keep FORS-M values near the "norm", which in this segment would appear to be about -25 kcal/mol. In this circumstance, positive FORS-D values (Figs. 3a, 4c) appear to partly countermand the high base-composition-driven stem-loop potential (Fig. 8d).
In a region where a strong positive Darwinian selection pressure on protein causing rapid sequence evolution would oppose the development of base order-dependent stem-loop potential (negative FORS-D values), the same positive selection pressure might also oppose the development of this countermanding (anti-stem-loop) base order-dependent stem-loop potential (positive FORS-D values). Thus, regions under positive Darwinian selection pressure should have FORS-D values fluctuating close to zero. The fact that FORS-D values significantly below the abscissa are observed in genes under strong positive selection (Forsdyke, 1995b, d;1996b), implies a hierarchy of power (anti-stem-loop potential > protein potential > stem-loop potential).
A possible explanation for the over-riding power of positive FORS-D values (below the abscissa) derives from the proposed role in recombination, or "accounting", of stem-loop structures extruded from duplex DNA (Bell & Forsdyke, unpublished work). The proposed "kissing" interactions between the tips of loops required for intra- or inter-chromosomal recombination or "accounting" would seem to require a very precise, highly reproducible, stem-loop architecture (Forsdyke, 1996a). This architecture would critically depend on the number and location of sites where the extrusion process could initiate (Murchie et al., 1992; Sinden, 1994). Extrusion would be difficult in regions of high base composition-dependent stem-loop potential (e.g. the CpG island of Figure 3), simply because the high (C+G)% favours retention of the duplex conformation. A countermanding base order-dependent stem-loop potential (positive FORS-D values) might be more compelling than the external force driving positive Darwinian selection.
10. Identification of genes in uncharted sequences
Although it is apparent that our understanding of the nature and relative strengths of the various pressures moulding the evolution of stem-loop potential is incomplete, one possible practical benefit of the present approach is apparent from data on the G0S2 gene (Fig. 3) and on the G0S3 gene (Heximer et al., 1996). The decline in base order-dependent stem-loop potential (FORS-D values below the abscissa) is most marked in the region of genes, relative to the flanking intergenic DNA. Thus folding of randomized sequence difference analysis (FORS-D analysis) may supplement existing methods of identifying genes in the numerous long uncharted DNA sequences which are accumulating in databases as part of various genome projects (Snyder & Stormo, 1995). It is shown elsewhere that analysis of Chargaff differences also has potential in this respect (Bell and Forsdyke, 1999a,b; Dang et al. 1998).
I thank L. Biswas and L. Russell for technical help, A. Cristillo and J. Gerlach for assistance with computer configuration, and T. Smith for advice on statistics. The work was supported by a grant from the Medical Research Council of Canada.
REFERENCES
BORER, P. N., DENGLER, B. & TINOCO, I. (1974). Stability of ribonucleic acid double-stranded helices. J. Mol. Biol. 86, 843-853.
BRESLAUER, K. A., FRANK, R., BLOCKER, H. & MARKY, L. A. (1986). Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 83, 3746- 3750.
CHEN, J-H., LE, S-Y., SHAPIRO, B., CURREY, K. M. & MAIZEL, J. V. (1990). A computational procedure for assessing the significance of RNA secondary structure. CABIOS 6, 7-18.
CRISTILLO, A. D., HEXIMER, S. P., RUSSELL, L. & FORSDYKE, D. R. (1997). Cyclosporin A inhibits early mRNA expression of G0 /G1 Switch Gene 2 (G0S2) in cultured human blood mononuclear cells. DNA Cell Biol. 16, 1449-1458.
CROSS, S. H. & BIRD, A. P. (1995). CpG islands and genes. Curr. Opin. Genet. Devel. 5, 309-314.
DANG, K. D., DUTT, P. B. & FORSDYKE, D. R. (1998). Chargaff difference analysis of the bithorax complex of Drosophila melanogaster. Biochem. Cell Biol. 76, 129-137.
FONTANA, W., KONINGS, D. A. M., STADLER, P. F. & SCHUSTER, P. (1993). Statistics of RNA secondary structures. Biopolymers 33, 1389-1404.
FORSDYKE, D. R. (1995a). A stem-loop "kissing" model for the initiation of recombination and the origin of introns. Mol. Biol. Evol. 12, 949-958.
FORSDYKE, D. R. (1995b). Conservation of stem-loop potential in introns of snake venom phospholipase A2 genes. An application of FORS-D analysis. Mol. Biol. Evol. 12, 1157-1165.
FORSDYKE, D. R. (1995c). Relative roles of primary sequence and (G+C)% in determining the hierarchy of frequencies of complementary trinucleotide pairs in DNAs of different species. J. Mol. Evol. 41, 573-581.
FORSDYKE, D. R. (1995d). Reciprocal relationship between stem-loop potential and substitution density in retroviral quasispecies under positive Darwinian selection. J. Mol. Evol. 41, 1022-1037.
FORSDYKE, D. R. (1996a). Different biological species "broadcast" their DNAs at different (G+C)% "wavelengths". J. theor. Biol. 178, 405-417.
FORSDYKE, D. R. (1996b). Stem-loop potential: a new way of evaluating positive Darwinian selection? Immunogenetics 43, 182-189.
GACY, A. M., GOELLNER, G., JURANIC, N., MACURA, S. & McMURRAY, C. T. (1995). Cell 81, 533-540. Trinucleotide repeats that expand in human disease form hairpin structures in vitro.
HEXIMER, S. P., CRISTILLO, A. D., RUSSELL, L., & FORSDYKE, D. R. (1996). Sequence analysis and expression in cultured lymphocytes of the human FOSB gene (G0S3). DNA Cell Biol. 15, 1025-1038.
KAGAWA, Y., NOJIMA, H., NUKIWA, N., ISHIZUKA, M., NAKAJIMA, T., YASUHARA, Y., et al. (1984). High G + C content in the third letter of codons of an extreme thermophile. J. Biol. Chem. 259, 2956-2960.
KARLIN, S., & BRENDEL, V. (1992). Chance and statistical significance in protein and DNA sequence analysis. Science 257, 39-49.
KLECKNER, N. & WEINER, B. M. (1993). Potential advantages of unstable interactions for pairing of chromosomes in meiotic, somatic and premeiotic cells. Cold Spring Harbour Symp. Quant. Biol. 58, 553-565.
KLECKNER, N. (1997). Interactions between and along chromosomes during meiosis. Harvey Lectures 91, 21-45.
LE, S-Y., CHEN, J-H. & MAIZEL, J. V. (1991). Detection of unusual RNA folding regions in HIV and SIV sequences. CABIOS 7, 51-55.
LE, S-Y. & MAIZEL, J. V. (1989). A method for assessing the statistical significance of RNA folding. J. theor. Biol. 138, 495-510.
LEWIS, E. B. (1954). The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 88, 225-239.
MURCHIE, A. I. H., BOWATER, R., ABOUL-ELA, F. & LILLEY, D. M. J. (1992). Helix opening transitions in supercoiled DNA. Biochem. Biophys. Acta 1131, 1-15.
NICHOLS, B. P., BLUMENBERG, M. & YANOFSKY, C. (1981). Comparison of the nucleotide sequence of trpA and sequences immediately beyond the trp operon of Klebsiella aerogenes, Salmonella typhi, and Escherichia coli. Nucleic Acids Res. 9, 1743- 1755.
NIELSEN, D. A., NOVORADOVSKY, A. & GOLDMAN, D. (1995). SSCP primer design based on single-strand DNA structure predicted by a DNA folding program. Nucleic Acids Res. 23, 2287-2291.
NUSSINOV. R. (1981). Eukaryotic dinucleotide preference rules and their implications for degenerate codon usage. J. Mol. Biol. 149, 125-131.
PRABHU, V. V. (1993). Symmetry observations in long nucleotide sequences. Nucleic Acids Res. 21, 2797-2800.
RADMAN, M., WAGNER, R. & KRICKER, M. C. (1993). Homologous DNA interactions in the evolution of gene and chromosome structure. Genome Anal. 7, 139- 154.
RUSSELL, L. & FORSDYKE, D. R. (1991). A human putative lymphocyte G0/G1 switch gene containing a CpG-rich island encodes a small basic protein with the potential to be phosphorylated. DNA Cell. Biol. 10, 581-591.
RYAN, B. F. & JOINER, B. L. (1994). Minitab Handbook. 3rd edition. Wadsworth Publishing, Belmont, California.
SANTALUCIA, J., ALLAWI, H. T. & SENEVIRATNE, P. A. (1996). Improved nearest neighbour parameters for predicting DNA duplex stability. Biochemistry 35, 3555-3562.
SHEN, L. X., CAI, Z. & TINOCO, I. (1995). RNA structure at high resolution. FASEB. J. 9, 1023-1033.
SINDEN, R. (1994). DNA Structure and Function. Academic Press, San Diego.
SMITHIES, O., ENGELS, W. R., DEVEREUX, J. R., SLIGHTOM, J. L. & SHEN, S. (1981). Base substitutions, length differences and DNA strand asymmetries in the human Gl and Al fetal globin gene region. Cell 26, 345-353.
SNYDER, E. E. & STORMO, G. D. (1995). Identification of protein-coding regions in genomic DNA. J. Mol. Biol. 240, 1-10.
TINOCO, I., UHLENBECK, O. C. & LEVINE, M. D. (1971). Estimating secondary structure in ribonucleic acids. Nature 230, 362-367.
TURNER, D. H., SUGIMOTO, N. & FREIER, S. M. (1988). RNA structure prediction. Annu. Rev. Biophys. Chem. 17, 167-192.
WELLS, R. D. (1996). Molecular basis of genetic instability of triplet repeats. J. Biol. Chem. 271, 2875-2878.
WU, C. -t. (1993). Transvection, nuclear structure and chromatin proteins. J. Cell Biol. 120, 587-590.
ZUKER, M. (1994). Prediction of RNA secondary structure by energy minimization. Meth. Molec. Biol. 25, 267-294.
ZUKER. M., JAEGER, J. A. & TURNER, D. H. (1991). A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structure determined by phylogenetic comparison. Nucleic Acids Res. 19, 2707- 2714.
Go to: Accounting Units in DNA (Bell & Forsdyke 1999a) (Click Here)
Go to: Folding single-stranded nucleic acid: conceptual problems (Forsdyke 2007) (Click Here)
Return to: Bioinformatics Index (Click Here)
Return to: Evolution Index (Click Here)
Return to: HomePage (Click Here)
This page was established circa 1998 and was last edited on 26 Jan 2008 by Donald Forsdyke